IsomiRome analysis reveals dysregulation of 5’ isomiRs during SARS-CoV-2 infection
et al., Computational and Structural Biotechnology Journal, doi:10.1016/j.csbj.2025.10.012, Oct 2025
In vitro study showing that 5' isomiRs, particularly miR-4485-3p|+1, are significantly dysregulated during SARS-CoV-2 infection in human lung cells and may contribute to viral pathogenesis by repressing host cellular processes.
Lyu et al., 8 Oct 2025, China, peer-reviewed, 8 authors.
Contact: quanyiwang@cpu.edu.cn, cwang1971@cpu.edu.cn, haiyanghu@cpu.edu.cn.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
IsomiRome analysis reveals dysregulation of 5’ isomiRs during SARS-CoV-2 infection
Computational and Structural Biotechnology Journal, doi:10.1016/j.csbj.2025.10.012
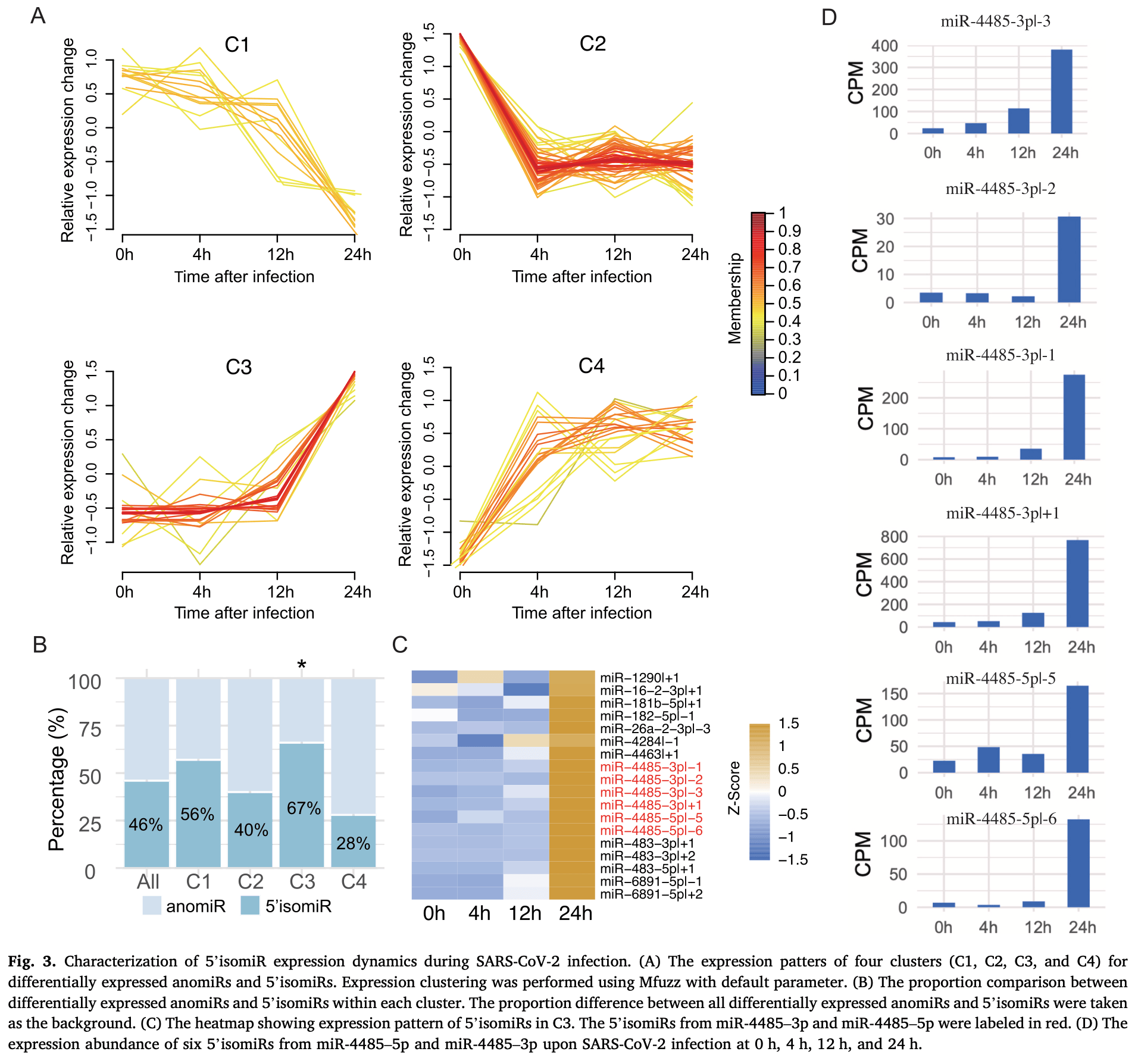
Annotated canonical miRNAs (anomiRs) are critical regulators of COVID-19 pathogenesis. Their 5′ isomiR counterparts, which harbor a shifted 5' end and an altered seed region relative to cognate anomiRs, represent an essential yet understudied component of the miRNAome. Herein, we performed an integrative analysis to investigate the expression and regulation of 5' isomiRs in SARS-CoV-2-infected human lung cells. Using Argonaute (AGO) immunoprecipitated small-RNA sequencing data, we established a repertoire of AGO-loaded 5' isomiRome encompassing 826 5' isomiRs from SARS-CoV-2-infected lung cells. A total of 54 5' isomiRs were differentially expressed, with the majority showing dysregulation 24 h after SARS-CoV-2 infection. Compared to anomiRs, 5' isomiRs were preferentially induced at 24 h post SARS-CoV-2 infection, including six 5' isomiRs derived from miR-4485 precursor. By focusing on a 5' isomiR of miR-4485-3p exhibiting one nucleotide downstream shift of the 5' end (miR-4485-3p|+1), we demonstrated that miR-4485-3p|+ 1 overexpression significantly repressed genes involved in cellular processes relating to cell cycle, cytoplasmic translation, and carbohydrate derivative metabolic process. Importantly, transcriptome integration further revealed that these cellular processes repressed by miR-4485-3p|+ 1 overexpression were strongly downregulated at 24 h following SARS-CoV-2 infection. Moreover, 26 out of 43 (60.5 %) hub genes co-repressed by miR-4485-3p|+ 1 and SARS-CoV-2 infection in these three cellular processes have reported association with COVID-19 in the literature. Taken together, our findings reveal the repertoire and temporal dynamics of AGO-loaded 5' isomiRs in SARS-CoV-2-infected lung cells and identify miR-4485-3p|+ 1 as a potential regulator of host pathways relevant to COVID-19 pathogenesis.
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supporting information Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2025.10.012.
References
Aiso, Ueda, 5'-isomiR is the most abundant sequence of miR-1246, a candidate biomarker of lung cancer, in serum, Mol Med Rep
Bartel, MicroRNAs: target recognition and regulatory functions, Cell
Bofill-De Ros, Luke, Guthridge, Mudunuri, Loss et al., Tumor IsomiR encyclopedia (TIE): a pan-cancer database of miRNA isoforms, Bioinformatics
Bolger, Lohse, Usadel, Trimmomatic: a flexible trimmer for illumina sequence data, Bioinformatics
Bouhaddou, Memon, Meyer, White, Rezelj et al., The global phosphorylation landscape of SARS-CoV-2 infection, Cell
Chen, Wang, Shyr, Liu, FindAdapt: a python package for fast and accurate adapter detection in small RNA sequencing, PLoS Comput Biol
Chen, Wu, He, Jiang, He, Metabolic alterations upon SARS-CoV-2 infection and potential therapeutic targets against coronavirus infection, Signal Transduct Target Ther
De Hoon, Taft, Hashimoto, Kanamori-Katayama, Kawaji et al., Cross-mapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries, Genome Res
Diallo, Jacob, Vion, Kozak, Mossman et al., Altered microRNA transcriptome in cultured human airway cells upon infection with SARS-CoV-2, Viruses
Fernandez-Valverde, Taft, Mattick, Dynamic isomir regulation in drosophila development, RNA
Gassen, Papies, Bajaj, Emanuel, Dethloff et al., SARS-CoV-2mediated dysregulation of metabolism and autophagy uncovers host-targeting antivirals, Nat Commun
Harbach, Tran, Kastrappis, Tran, Grimley et al., Polarized Calu-3 cells serve as an intermediary model for SARS-CoV-2 infection, Methods Mol Biol
Henzinger, Barth, Klec, Pichler, Non-Coding RNAs and SARS-Related coronaviruses, Viruses
Hong, Hu, Zhang, Liu, Yan et al., Integrative analysis identifies region-and sex-specific gene networks and Mef2c as a mediator of anxiety-like behavior, Cell Rep
Hu, Guo, Xi, Yan, Fu et al., MicroRNA expression and regulation in human, chimpanzee, and macaque brains, PLoS Genet
Hu, Guo, Zhou, Shi, Characteristics of SARS-CoV-2 and COVID-19, Nat Rev Microbiol
Hu, He, Fominykh, Yan, Guo et al., Evolution of the humanspecific microRNA miR-941, Nat Commun
Hu, Liu, Hu, Jiang, Yang et al., Recently evolved tumor suppressor transcript TP73-AS1 functions as sponge of Human-Specific miR-941, Mol Biol Evol
Jouravleva, Golovenko, Demo, Dutcher, Hall et al., Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB, Mol Cell
Kaur, Kapila, Kapila, Kumar, Upadhyay et al., Tmprss2 specific miRNAs as promising regulators for SARS-CoV-2 entry checkpoint, Virus Res
Kim, Paggi, Park, Bennett, Salzberg, Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype, Nat Biotechnol
Kozomara, Birgaoanu, Griffiths-Jones, miRBase: from microRNA sequences to function, Nucleic Acids Res
Krishnan, Nordqvist, Ambikan, Gupta, Sperk et al., Metabolic perturbation associated with COVID-19 disease severity and SARS-CoV-2 replication, Mol Cell Proteom
Kumar, Ef, mfuzz: a software package for soft clustering of microarray data, Bioinformation
Lambert, Lambert, Clarke, Hooper, Porter et al., Angiotensin-converting enzyme 2 is subject to post-transcriptional regulation by miR-421, Clin Sci (Lond)
Lamers, Haagmans, SARS-CoV-2 pathogenesis, Nat Rev Microbiol
Langmead, Trapnell, Pop, Salzberg, Ultrafast and memory-efficient alignment of short DNA sequences to the human genome, Genome Biol
Li, Michels, Tosun, Jung, Kappes et al., 5'isomiR-183-5p|+2 elicits tumor suppressor activity in a negative feedback loop with E2F1, J Exp Clin Cancer Res
Liao, Smyth, Shi, Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features, Bioinformatics
Ma, Wu, Niu, Zhang, Jia et al., A sliding-bulge structure at the dicer processing site of pre-miRNAs regulates alternative dicer processing to generate 5′-isomiRs, Heliyon
Mahlab-Aviv, Linial, Linial, A cell-based probabilistic approach unveils the concerted action of miRNAs, PLoS Comput Biol
Maimaiti, Li, Cheng, Zhong, Hu et al., Blocking cGAS-STING pathway promotes post-stroke functional recovery in an extended treatment window via facilitating remyelination, Med
Markov, Ghafari, Beer, Lythgoe, Simmonds et al., The evolution of SARS-CoV-2, Nat Rev Microbiol
Marti, Pantano, Banez-Coronel, Llorens, Minones-Moyano et al., A myriad of miRNA variants in control and huntington's disease brain regions detected by massively parallel sequencing, Nucleic Acids Res
Neilsen, Goodall, Bracken, IsomiRs-the overlooked repertoire in the dynamic microRNAome, Trends Genet
Park, Yang, Seo, Nam, Nam, hnRNPC induces isoform shifts in miR-21-5p leading to cancer development, Exp Mol Med
Pawlica, Yario, White, Wang, Moss et al., SARS-CoV-2 expresses a microRNA-like small RNA able to selectively repress host genes, Proc Natl Acad Sci
Robinson, Mccarthy, Smyth, Edger: a bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics
Song, Zhang, Li, Maimaiti, Sun et al., m(6)A-mediated modulation coupled with transcriptional regulation shapes long noncoding RNA repertoire of the cGAS-STING signaling, Comput Struct Biotechnol J
Su, Chen, Qi, Shi, Feng et al., A Mini-Review on cell cycle regulation of coronavirus infection, Front Vet Sci
Sui, Li, Zhao, Zhao, Hao et al., Host cell cycle checkpoint as antiviral target for SARS-CoV-2 revealed by integrative transcriptome and proteome analyses, Signal Transduct Target Ther
Szklarczyk, Kirsch, Koutrouli, Nastou, Mehryary et al., The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest, Nucleic Acids Res
Tan, Chan, Molnar, Sarkar, Alexieva et al., 5' isomir variation is of functional and evolutionary importance, Nucleic Acids Res
Tang, Luo, Cui, Ni, Yuan et al., MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins, Arthritis Rheum
Tu, Yu, Hua, Li, Liu et al., Combinatorial network of primary and secondary microRNA-driven regulatory mechanisms, Nucleic Acids Res
Van Der Kwast, Woudenberg, Quax, Nossent, MicroRNA-411 and its 5′-IsomiR have distinct targets and functions and are differentially regulated in the vasculature under ischemia, Mol Ther
Vora, Fontana, Mao, Leger, Zhang et al., Targeting stem-loop 1 of the SARS-CoV-2 5′ UTR to suppress viral translation and Nsp1 evasion, Proc Natl Acad Sci
Wagner, Meese, Keller, The intricacies of isomirs: from classification to clinical relevance, Trends Genet
Wyler, Mosbauer, Franke, Diag, Gottula et al., Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy, iScience
Yu, Pillman, Neilsen, Toubia, Lawrence et al., Naturally existing isoforms of miR-222 have distinct functions, Nucleic Acids Res
Zhang, Zhu, Wang, Li, Gao, Translational control of COVID-19 and its therapeutic implication, Front Immunol
DOI record:
{
"DOI": "10.1016/j.csbj.2025.10.012",
"ISSN": [
"2001-0370"
],
"URL": "http://dx.doi.org/10.1016/j.csbj.2025.10.012",
"alternative-id": [
"S2001037025004143"
],
"assertion": [
{
"label": "This article is maintained by",
"name": "publisher",
"value": "Elsevier"
},
{
"label": "Article Title",
"name": "articletitle",
"value": "IsomiRome analysis reveals dysregulation of 5’ isomiRs during SARS-CoV-2 infection"
},
{
"label": "Journal Title",
"name": "journaltitle",
"value": "Computational and Structural Biotechnology Journal"
},
{
"label": "CrossRef DOI link to publisher maintained version",
"name": "articlelink",
"value": "https://doi.org/10.1016/j.csbj.2025.10.012"
},
{
"label": "Content Type",
"name": "content_type",
"value": "article"
},
{
"label": "Copyright",
"name": "copyright",
"value": "© 2025 The Authors. Published by Elsevier B.V. on behalf of Research Network of Computational and Structural Biotechnology."
}
],
"author": [
{
"affiliation": [],
"family": "Lyu",
"given": "Ming-Ju Amy",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0000-0003-4920-6141",
"affiliation": [],
"authenticated-orcid": false,
"family": "Maimaiti",
"given": "Munire",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0009-0006-2573-7086",
"affiliation": [],
"authenticated-orcid": false,
"family": "Zhong",
"given": "Ziwei",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hu",
"given": "Jiameng",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zhang",
"given": "Xiang",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-6773-0057",
"affiliation": [],
"authenticated-orcid": false,
"family": "Wang",
"given": "Quanyi",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wang",
"given": "Chen",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-5348-5641",
"affiliation": [],
"authenticated-orcid": false,
"family": "Hu",
"given": "Haiyang",
"sequence": "additional"
}
],
"container-title": "Computational and Structural Biotechnology Journal",
"container-title-short": "Computational and Structural Biotechnology Journal",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"csbj.org",
"elsevier.com",
"sciencedirect.com"
]
},
"created": {
"date-parts": [
[
2025,
10,
9
]
],
"date-time": "2025-10-09T07:59:39Z",
"timestamp": 1759996779000
},
"deposited": {
"date-parts": [
[
2025,
10,
29
]
],
"date-time": "2025-10-29T15:05:11Z",
"timestamp": 1761750311000
},
"funder": [
{
"DOI": "10.13039/501100001809",
"award": [
"82471347"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100001809",
"id-type": "DOI"
}
],
"name": "National Natural Science Foundation of China"
},
{
"DOI": "10.13039/501100002857",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100002857",
"id-type": "DOI"
}
],
"name": "China Pharmaceutical University"
},
{
"DOI": "10.13039/501100012166",
"award": [
"2021YFF0702003",
"2022YFC2303200"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100012166",
"id-type": "DOI"
}
],
"name": "National Key Research and Development Program of China"
}
],
"indexed": {
"date-parts": [
[
2025,
10,
29
]
],
"date-time": "2025-10-29T15:09:31Z",
"timestamp": 1761750571759,
"version": "build-2065373602"
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2025
]
]
},
"language": "en",
"license": [
{
"URL": "https://www.elsevier.com/tdm/userlicense/1.0/",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
1,
1
]
],
"date-time": "2025-01-01T00:00:00Z",
"timestamp": 1735689600000
}
},
{
"URL": "https://www.elsevier.com/legal/tdmrep-license",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
1,
1
]
],
"date-time": "2025-01-01T00:00:00Z",
"timestamp": 1735689600000
}
},
{
"URL": "http://creativecommons.org/licenses/by-nc-nd/4.0/",
"content-version": "vor",
"delay-in-days": 279,
"start": {
"date-parts": [
[
2025,
10,
7
]
],
"date-time": "2025-10-07T00:00:00Z",
"timestamp": 1759795200000
}
}
],
"link": [
{
"URL": "https://api.elsevier.com/content/article/PII:S2001037025004143?httpAccept=text/xml",
"content-type": "text/xml",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://api.elsevier.com/content/article/PII:S2001037025004143?httpAccept=text/plain",
"content-type": "text/plain",
"content-version": "vor",
"intended-application": "text-mining"
}
],
"member": "78",
"original-title": [],
"page": "4352-4362",
"prefix": "10.1016",
"published": {
"date-parts": [
[
2025
]
]
},
"published-print": {
"date-parts": [
[
2025
]
]
},
"publisher": "Elsevier BV",
"reference": [
{
"DOI": "10.1038/s41579-020-00459-7",
"article-title": "Characteristics of SARS-CoV-2 and COVID-19",
"author": "Hu",
"doi-asserted-by": "crossref",
"first-page": "141",
"issue": "3",
"journal-title": "Nat Rev Microbiol",
"key": "10.1016/j.csbj.2025.10.012_bib1",
"volume": "19",
"year": "2021"
},
{
"DOI": "10.1038/s41579-022-00713-0",
"article-title": "SARS-CoV-2 pathogenesis",
"author": "Lamers",
"doi-asserted-by": "crossref",
"first-page": "270",
"issue": "5",
"journal-title": "Nat Rev Microbiol",
"key": "10.1016/j.csbj.2025.10.012_bib2",
"volume": "20",
"year": "2022"
},
{
"DOI": "10.1073/pnas.2117198119",
"article-title": "Targeting stem-loop 1 of the SARS-CoV-2 5′ UTR to suppress viral translation and Nsp1 evasion",
"author": "Vora",
"doi-asserted-by": "crossref",
"issue": "9",
"journal-title": "Proc Natl Acad Sci USA",
"key": "10.1016/j.csbj.2025.10.012_bib3",
"volume": "119",
"year": "2022"
},
{
"DOI": "10.1038/s41392-022-01296-1",
"article-title": "Host cell cycle checkpoint as antiviral target for SARS-CoV-2 revealed by integrative transcriptome and proteome analyses",
"author": "Sui",
"doi-asserted-by": "crossref",
"first-page": "21",
"issue": "1",
"journal-title": "Signal Transduct Target Ther",
"key": "10.1016/j.csbj.2025.10.012_bib4",
"volume": "8",
"year": "2023"
},
{
"DOI": "10.3389/fvets.2020.586826",
"article-title": "A Mini-Review on cell cycle regulation of coronavirus infection",
"author": "Su",
"doi-asserted-by": "crossref",
"journal-title": "Front Vet Sci",
"key": "10.1016/j.csbj.2025.10.012_bib5",
"volume": "7",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.06.034",
"article-title": "The global phosphorylation landscape of SARS-CoV-2 infection",
"author": "Bouhaddou",
"doi-asserted-by": "crossref",
"first-page": "685",
"issue": "3",
"journal-title": "Cell",
"key": "10.1016/j.csbj.2025.10.012_bib6",
"volume": "182",
"year": "2020"
},
{
"article-title": "Translational control of COVID-19 and its therapeutic implication",
"author": "Zhang",
"journal-title": "Front Immunol",
"key": "10.1016/j.csbj.2025.10.012_bib7",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1038/s41392-023-01510-8",
"article-title": "Metabolic alterations upon SARS-CoV-2 infection and potential therapeutic targets against coronavirus infection",
"author": "Chen",
"doi-asserted-by": "crossref",
"first-page": "237",
"issue": "1",
"journal-title": "Signal Transduct Target Ther",
"key": "10.1016/j.csbj.2025.10.012_bib8",
"volume": "8",
"year": "2023"
},
{
"DOI": "10.1016/j.mcpro.2021.100159",
"article-title": "Metabolic perturbation associated with COVID-19 disease severity and SARS-CoV-2 replication",
"author": "Krishnan",
"doi-asserted-by": "crossref",
"journal-title": "Mol Cell Proteom",
"key": "10.1016/j.csbj.2025.10.012_bib9",
"volume": "20",
"year": "2021"
},
{
"DOI": "10.1038/s41467-021-24007-w",
"article-title": "SARS-CoV-2-mediated dysregulation of metabolism and autophagy uncovers host-targeting antivirals",
"author": "Gassen",
"doi-asserted-by": "crossref",
"first-page": "3818",
"issue": "1",
"journal-title": "Nat Commun",
"key": "10.1016/j.csbj.2025.10.012_bib10",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1038/s41579-023-00878-2",
"article-title": "The evolution of SARS-CoV-2",
"author": "Markov",
"doi-asserted-by": "crossref",
"first-page": "361",
"issue": "6",
"journal-title": "Nat Rev Microbiol",
"key": "10.1016/j.csbj.2025.10.012_bib11",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.1016/j.cell.2009.01.002",
"article-title": "MicroRNAs: target recognition and regulatory functions",
"author": "Bartel",
"doi-asserted-by": "crossref",
"first-page": "215",
"issue": "2",
"journal-title": "Cell",
"key": "10.1016/j.csbj.2025.10.012_bib12",
"volume": "136",
"year": "2009"
},
{
"DOI": "10.1093/molbev/msy022",
"article-title": "Recently evolved tumor suppressor transcript TP73-AS1 functions as sponge of Human-Specific miR-941",
"author": "Hu",
"doi-asserted-by": "crossref",
"first-page": "1063",
"issue": "5",
"journal-title": "Mol Biol Evol",
"key": "10.1016/j.csbj.2025.10.012_bib13",
"volume": "35",
"year": "2018"
},
{
"DOI": "10.1038/ncomms2146",
"article-title": "Evolution of the human-specific microRNA miR-941",
"author": "Hu",
"doi-asserted-by": "crossref",
"first-page": "1145",
"journal-title": "Nat Commun",
"key": "10.1016/j.csbj.2025.10.012_bib14",
"volume": "3",
"year": "2012"
},
{
"DOI": "10.1371/journal.pgen.1002327",
"article-title": "MicroRNA expression and regulation in human, chimpanzee, and macaque brains",
"author": "Hu",
"doi-asserted-by": "crossref",
"issue": "10",
"journal-title": "PLoS Genet",
"key": "10.1016/j.csbj.2025.10.012_bib15",
"volume": "7",
"year": "2011"
},
{
"DOI": "10.1002/art.24436",
"article-title": "MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins",
"author": "Tang",
"doi-asserted-by": "crossref",
"first-page": "1065",
"issue": "4",
"journal-title": "Arthritis Rheum",
"key": "10.1016/j.csbj.2025.10.012_bib16",
"volume": "60",
"year": "2009"
},
{
"DOI": "10.1093/nar/gky1141",
"article-title": "miRBase: from microRNA sequences to function",
"author": "Kozomara",
"doi-asserted-by": "crossref",
"first-page": "D155",
"issue": "D1",
"journal-title": "Nucleic Acids Res",
"key": "10.1016/j.csbj.2025.10.012_bib17",
"volume": "47",
"year": "2019"
},
{
"DOI": "10.1042/CS20130420",
"article-title": "Angiotensin-converting enzyme 2 is subject to post-transcriptional regulation by miR-421",
"author": "Lambert",
"doi-asserted-by": "crossref",
"first-page": "243",
"issue": "4",
"journal-title": "Clin Sci (Lond)",
"key": "10.1016/j.csbj.2025.10.012_bib18",
"volume": "127",
"year": "2014"
},
{
"DOI": "10.1016/j.virusres.2020.198275",
"article-title": "Tmprss2 specific miRNAs as promising regulators for SARS-CoV-2 entry checkpoint",
"author": "Kaur",
"doi-asserted-by": "crossref",
"journal-title": "Virus Res",
"key": "10.1016/j.csbj.2025.10.012_bib19",
"volume": "294",
"year": "2021"
},
{
"DOI": "10.3390/v12121374",
"article-title": "Non-Coding RNAs and SARS-Related coronaviruses",
"author": "Henzinger",
"doi-asserted-by": "crossref",
"issue": "12",
"journal-title": "Viruses",
"key": "10.1016/j.csbj.2025.10.012_bib20",
"volume": "12",
"year": "2020"
},
{
"DOI": "10.1016/j.tig.2024.05.007",
"article-title": "The intricacies of isomirs: from classification to clinical relevance",
"author": "Wagner",
"doi-asserted-by": "crossref",
"first-page": "784",
"issue": "9",
"journal-title": "Trends Genet",
"key": "10.1016/j.csbj.2025.10.012_bib21",
"volume": "40",
"year": "2024"
},
{
"DOI": "10.1093/nar/gku656",
"article-title": "5' isomir variation is of functional and evolutionary importance",
"author": "Tan",
"doi-asserted-by": "crossref",
"first-page": "9424",
"issue": "14",
"journal-title": "Nucleic Acids Res",
"key": "10.1016/j.csbj.2025.10.012_bib22",
"volume": "42",
"year": "2014"
},
{
"DOI": "10.1093/nar/gkq575",
"article-title": "A myriad of miRNA variants in control and huntington's disease brain regions detected by massively parallel sequencing",
"author": "Marti",
"doi-asserted-by": "crossref",
"first-page": "7219",
"issue": "20",
"journal-title": "Nucleic Acids Res",
"key": "10.1016/j.csbj.2025.10.012_bib23",
"volume": "38",
"year": "2010"
},
{
"DOI": "10.1261/rna.2379610",
"article-title": "Dynamic isomir regulation in drosophila development",
"author": "Fernandez-Valverde",
"doi-asserted-by": "crossref",
"first-page": "1881",
"issue": "10",
"journal-title": "RNA",
"key": "10.1016/j.csbj.2025.10.012_bib24",
"volume": "16",
"year": "2010"
},
{
"DOI": "10.1016/j.ymthe.2019.10.002",
"article-title": "MicroRNA-411 and its 5′-IsomiR have distinct targets and functions and are differentially regulated in the vasculature under ischemia",
"author": "van der Kwast",
"doi-asserted-by": "crossref",
"first-page": "157",
"issue": "1",
"journal-title": "Mol Ther",
"key": "10.1016/j.csbj.2025.10.012_bib25",
"volume": "28",
"year": "2020"
},
{
"DOI": "10.1016/j.tig.2012.07.005",
"article-title": "IsomiRs--the overlooked repertoire in the dynamic microRNAome",
"author": "Neilsen",
"doi-asserted-by": "crossref",
"first-page": "544",
"issue": "11",
"journal-title": "Trends Genet",
"key": "10.1016/j.csbj.2025.10.012_bib26",
"volume": "28",
"year": "2012"
},
{
"DOI": "10.1093/bioinformatics/btab172",
"article-title": "Tumor IsomiR encyclopedia (TIE): a pan-cancer database of miRNA isoforms",
"author": "Bofill-De Ros",
"doi-asserted-by": "crossref",
"first-page": "3023",
"issue": "18",
"journal-title": "Bioinformatics",
"key": "10.1016/j.csbj.2025.10.012_bib27",
"volume": "37",
"year": "2021"
},
{
"DOI": "10.1038/s12276-022-00792-2",
"article-title": "hnRNPC induces isoform shifts in miR-21-5p leading to cancer development",
"author": "Park",
"doi-asserted-by": "crossref",
"first-page": "812",
"issue": "6",
"journal-title": "Exp Mol Med",
"key": "10.1016/j.csbj.2025.10.012_bib28",
"volume": "54",
"year": "2022"
},
{
"DOI": "10.1093/nar/gkx788",
"article-title": "Naturally existing isoforms of miR-222 have distinct functions",
"author": "Yu",
"doi-asserted-by": "crossref",
"first-page": "11371",
"issue": "19",
"journal-title": "Nucleic Acids Res",
"key": "10.1016/j.csbj.2025.10.012_bib29",
"volume": "45",
"year": "2017"
},
{
"DOI": "10.1073/pnas.2116668118",
"article-title": "SARS-CoV-2 expresses a microRNA-like small RNA able to selectively repress host genes",
"author": "Pawlica",
"doi-asserted-by": "crossref",
"issue": "52",
"journal-title": "Proc Natl Acad Sci USA",
"key": "10.1016/j.csbj.2025.10.012_bib30",
"volume": "118",
"year": "2021"
},
{
"DOI": "10.1371/journal.pcbi.1011786",
"article-title": "FindAdapt: a python package for fast and accurate adapter detection in small RNA sequencing",
"author": "Chen",
"doi-asserted-by": "crossref",
"issue": "1",
"journal-title": "PLoS Comput Biol",
"key": "10.1016/j.csbj.2025.10.012_bib31",
"volume": "20",
"year": "2024"
},
{
"DOI": "10.1186/gb-2009-10-3-r25",
"article-title": "Ultrafast and memory-efficient alignment of short DNA sequences to the human genome",
"author": "Langmead",
"doi-asserted-by": "crossref",
"issue": "3",
"journal-title": "Genome Biol",
"key": "10.1016/j.csbj.2025.10.012_bib32",
"volume": "10",
"year": "2009"
},
{
"DOI": "10.1101/gr.095273.109",
"article-title": "Cross-mapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries",
"author": "de Hoon",
"doi-asserted-by": "crossref",
"first-page": "257",
"issue": "2",
"journal-title": "Genome Res",
"key": "10.1016/j.csbj.2025.10.012_bib33",
"volume": "20",
"year": "2010"
},
{
"DOI": "10.1016/j.isci.2021.102151",
"article-title": "Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy",
"author": "Wyler",
"doi-asserted-by": "crossref",
"issue": "3",
"journal-title": "iScience",
"key": "10.1016/j.csbj.2025.10.012_bib34",
"volume": "24",
"year": "2021"
},
{
"DOI": "10.1093/bioinformatics/btp616",
"article-title": "Edger: a bioconductor package for differential expression analysis of digital gene expression data",
"author": "Robinson",
"doi-asserted-by": "crossref",
"first-page": "139",
"issue": "1",
"journal-title": "Bioinformatics",
"key": "10.1016/j.csbj.2025.10.012_bib35",
"volume": "26",
"year": "2010"
},
{
"DOI": "10.3390/v15020496",
"article-title": "Altered microRNA transcriptome in cultured human airway cells upon infection with SARS-CoV-2",
"author": "Diallo",
"doi-asserted-by": "crossref",
"issue": "2",
"journal-title": "Viruses",
"key": "10.1016/j.csbj.2025.10.012_bib36",
"volume": "15",
"year": "2023"
},
{
"DOI": "10.6026/97320630002005",
"article-title": "M EF: mfuzz: a software package for soft clustering of microarray data",
"author": "Kumar",
"doi-asserted-by": "crossref",
"first-page": "5",
"issue": "1",
"journal-title": "Bioinformation",
"key": "10.1016/j.csbj.2025.10.012_bib37",
"volume": "2",
"year": "2007"
},
{
"DOI": "10.1016/j.medj.2024.03.018",
"article-title": "Blocking cGAS-STING pathway promotes post-stroke functional recovery in an extended treatment window via facilitating remyelination",
"author": "Maimaiti",
"doi-asserted-by": "crossref",
"first-page": "622",
"issue": "6",
"journal-title": "Med",
"key": "10.1016/j.csbj.2025.10.012_bib38",
"volume": "5",
"year": "2024"
},
{
"DOI": "10.1016/j.csbj.2022.04.002",
"article-title": "m(6)A-mediated modulation coupled with transcriptional regulation shapes long noncoding RNA repertoire of the cGAS-STING signaling",
"author": "Song",
"doi-asserted-by": "crossref",
"first-page": "1785",
"journal-title": "Comput Struct Biotechnol J",
"key": "10.1016/j.csbj.2025.10.012_bib39",
"volume": "20",
"year": "2022"
},
{
"DOI": "10.1016/j.celrep.2024.114455",
"article-title": "Integrative analysis identifies region- and sex-specific gene networks and Mef2c as a mediator of anxiety-like behavior",
"author": "Hong",
"doi-asserted-by": "crossref",
"issue": "7",
"journal-title": "Cell Rep",
"key": "10.1016/j.csbj.2025.10.012_bib40",
"volume": "43",
"year": "2024"
},
{
"DOI": "10.1093/bioinformatics/btu170",
"article-title": "Trimmomatic: a flexible trimmer for illumina sequence data",
"author": "Bolger",
"doi-asserted-by": "crossref",
"first-page": "2114",
"issue": "15",
"journal-title": "Bioinformatics",
"key": "10.1016/j.csbj.2025.10.012_bib41",
"volume": "30",
"year": "2014"
},
{
"DOI": "10.1038/s41587-019-0201-4",
"article-title": "Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype",
"author": "Kim",
"doi-asserted-by": "crossref",
"first-page": "907",
"issue": "8",
"journal-title": "Nat Biotechnol",
"key": "10.1016/j.csbj.2025.10.012_bib42",
"volume": "37",
"year": "2019"
},
{
"DOI": "10.1093/bioinformatics/btt656",
"article-title": "Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features",
"author": "Liao",
"doi-asserted-by": "crossref",
"first-page": "923",
"issue": "7",
"journal-title": "Bioinformatics",
"key": "10.1016/j.csbj.2025.10.012_bib43",
"volume": "30",
"year": "2014"
},
{
"DOI": "10.1007/7651_2024_602",
"article-title": "Polarized Calu-3 cells serve as an intermediary model for SARS-CoV-2 infection",
"author": "Harbach",
"doi-asserted-by": "crossref",
"journal-title": "Methods Mol Biol",
"key": "10.1016/j.csbj.2025.10.012_bib44",
"year": "2025"
},
{
"DOI": "10.1371/journal.pcbi.1007204",
"article-title": "A cell-based probabilistic approach unveils the concerted action of miRNAs",
"author": "Mahlab-Aviv",
"doi-asserted-by": "crossref",
"issue": "12",
"journal-title": "PLoS Comput Biol",
"key": "10.1016/j.csbj.2025.10.012_bib45",
"volume": "15",
"year": "2019"
},
{
"DOI": "10.1093/nar/gkp638",
"article-title": "Combinatorial network of primary and secondary microRNA-driven regulatory mechanisms",
"author": "Tu",
"doi-asserted-by": "crossref",
"first-page": "5969",
"issue": "18",
"journal-title": "Nucleic Acids Res",
"key": "10.1016/j.csbj.2025.10.012_bib46",
"volume": "37",
"year": "2009"
},
{
"DOI": "10.1093/nar/gkac1000",
"article-title": "The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest",
"author": "Szklarczyk",
"doi-asserted-by": "crossref",
"first-page": "D638",
"issue": "D1",
"journal-title": "Nucleic Acids Res",
"key": "10.1016/j.csbj.2025.10.012_bib47",
"volume": "51",
"year": "2023"
},
{
"DOI": "10.3892/mmr.2023.12979",
"article-title": "5'‑isomiR is the most abundant sequence of miR‑1246, a candidate biomarker of lung cancer, in serum",
"author": "Aiso",
"doi-asserted-by": "crossref",
"issue": "4",
"journal-title": "Mol Med Rep",
"key": "10.1016/j.csbj.2025.10.012_bib48",
"volume": "27",
"year": "2023"
},
{
"DOI": "10.1186/s13046-022-02380-8",
"article-title": "5'isomiR-183-5p|+2 elicits tumor suppressor activity in a negative feedback loop with E2F1",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "190",
"issue": "1",
"journal-title": "J Exp Clin Cancer Res",
"key": "10.1016/j.csbj.2025.10.012_bib49",
"volume": "41",
"year": "2022"
},
{
"DOI": "10.1016/j.heliyon.2016.e00148",
"article-title": "A sliding-bulge structure at the dicer processing site of pre-miRNAs regulates alternative dicer processing to generate 5′-isomiRs",
"author": "Ma",
"doi-asserted-by": "crossref",
"issue": "9",
"journal-title": "Heliyon",
"key": "10.1016/j.csbj.2025.10.012_bib50",
"volume": "2",
"year": "2016"
},
{
"DOI": "10.1016/j.molcel.2022.09.002",
"article-title": "Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB",
"author": "Jouravleva",
"doi-asserted-by": "crossref",
"first-page": "4049",
"issue": "21",
"journal-title": "Mol Cell",
"key": "10.1016/j.csbj.2025.10.012_bib51",
"volume": "82",
"year": "2022"
}
],
"reference-count": 51,
"references-count": 51,
"relation": {},
"resource": {
"primary": {
"URL": "https://linkinghub.elsevier.com/retrieve/pii/S2001037025004143"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"special_numbering": "C",
"subject": [],
"subtitle": [],
"title": "IsomiRome analysis reveals dysregulation of 5’ isomiRs during SARS-CoV-2 infection",
"type": "journal-article",
"update-policy": "https://doi.org/10.1016/elsevier_cm_policy",
"volume": "27"
}