Host proviral and antiviral factors for SARS-CoV-2
et al., Virus Genes, doi:10.1007/s11262-021-01869-2, Sep 2021
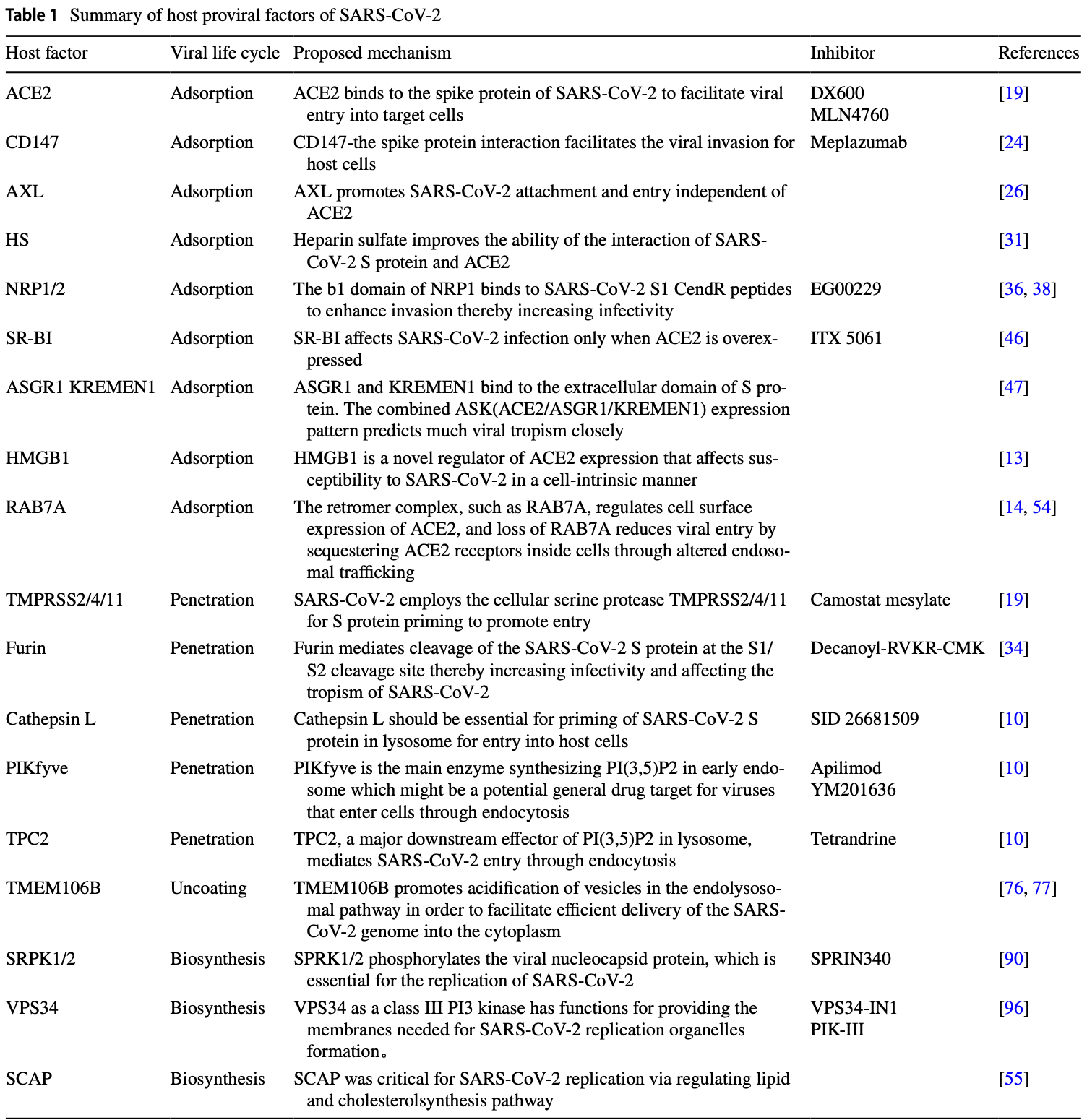
Review of host proviral and antiviral factors for SARS-CoV-2. Authors summarize current understanding of the interplay between SARS-CoV-2 and host cellular factors during virus entry and replication. They highlight ACE2 as the main receptor for viral entry, aided by cofactors like CD147, heparan sulfate, and neuropilin-1/2 to facilitate spike protein binding. Intracellular proteases like TMPRSS2 prime the spike protein to enable membrane fusion. Following entry and uncoating, cytosolic factors like SRPK1/2 kinases and VPS34 lipid kinase promote viral replication complex assembly and genome replication. Antiviral factors include defensin HD5 that blocks ACE2, cholesterol 25-hydroxylase that restricts membrane fusion, and ZAP/LARP1 that inhibit viral RNA production.
Lv et al., 11 Sep 2021, peer-reviewed, 2 authors.
Contact: armzhang@hotmail.com.
Host proviral and antiviral factors for SARS-CoV-2
Virus Genes, doi:10.1007/s11262-021-01869-2
Throughout the viral life cycle, interplays between cellular host factors and virus determine the infectious capacity of the virus. The pandemic of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) poses a great threat to human life and health. Extensive studies identified a number of host proviral and antiviral factors for SARS-CoV-2. In this review, we summarize the current understanding of the interplay between SARS-CoV-2 and cellular factors during virus entry and replication. Our review will highlight the future direction of study on the infection and pathogenesis of SARS-CoV-2, as well as novel therapeutic strategies and effective antiviral targets for COVID-19.
Author contributions LL wrote the manuscript. LZ conceived the work and revised the manuscript. Both authors read and approved the final manuscript.
Conflict of interest The authors declare no conflict of interest.
Informed consent Not applicable. Research involving human participants and/or animals Not applicable.
References
Abrams, Johnson, Perelman, Zhang, Endapally et al., Oxysterols provide innate immunity to bacterial infection by mobilizing cell surface accessible cholesterol, Nat Microbiol
Acton, Rigotti, Landschulz, Xu, Hobbs et al., Identification of scavenger receptor SR-BI as a high density lipoprotein receptor, Science
Andersson, Ottestad, Tracey, Extracellular HMGB1: a therapeutic target in severe pulmonary inflammation including COVID-19?, Mol Med
Andersson, Yang, Harris, High-mobility group box 1 protein (HMGB1) operates as an alarmin outside as well as inside cells, Semin Immunol
Aubol, Jamros, Mcglone, Adams, Splicing kinase SRPK1 conforms to the landscape of its SR protein substrate, Biochemistry
Backer, The intricate regulation and complex functions of the Class III phosphoinositide 3-kinase Vps34, Biochem J
Baggen, Persoons, Vanstreels, Jansen, Van Looveren et al., Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2, Nat Genet
Bajimaya, Frankl, Hayashi, Takimoto, Cholesterol is required for stability and infectivity of influenza A and respiratory syncytial viruses, Virology
Beeraka, Sadhu, Madhunapantula, Pragada, Svistunov et al., Strategies for targeting SARS CoV-2: small molecule inhibitors-the current status, Front Immunol
Bouhaddou, Memon, Meyer, White, Rezelj et al., The global phosphorylation landscape of SARS-CoV-2 infection, Cell
Brady, Zheng, Murphy, Huang, Hu, The frontotemporal lobar degeneration risk factor, TMEM106B, regulates lysosomal morphology and function, Hum Mol Genet
Braun, Sauter, Furin-mediated protein processing in infectious diseases and cancer, Clin Transl Immunol
Cagno, Tseligka, Jones, Tapparel, Heparan sulfate proteoglycans and viral attachment: true receptors or adaptation bias?, Viruses
Calcaterra, Armas, Weiner, Borgognone, CNBP: a multifunctional nucleic acid chaperone involved in cell death and proliferation control, IUBMB Life
Cantuti-Castelvetri, Ojha, Pedro, Djannatian, Franz et al., Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity, Science
Carette, Raaben, Wong, Herbert, Obernosterer et al., Ebola virus entry requires the cholesterol transporter Niemann-Pick C1, Nature
Catanese, Ansuini, Graziani, Huby, Moreau et al., Role of scavenger receptor class B type I in hepatitis C virus entry: kinetics and molecular determinants, J Virol
Chen, Mi, Xu, Yu, Wang et al., Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus, J Infect Dis
Cheng, Chao, Li, Chiu, Kao et al., Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects, Cell Rep
Chu, Hu, Huang, Chai, Zhou et al., Host and viral determinants for efficient SARS-CoV-2 infection of the human lung, Nat Commun
Chua, Lukassen, Trump, Hennig, Wendisch et al., COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis, Nat Biotechnol
Clausen, Sandoval, Spliid, Pihl, Perrett et al., SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2, Cell
Crosnier, Bustamante, Bartholdson, Bei, Theron et al., Basigin is a receptor essential for erythrocyte invasion by plasmodium falciparum, Nature
Daly, Simonetti, Klein, Chen, Williamson et al., Neuropilin-1 is a host factor for SARS-CoV-2 infection, Science
Daub, Blencke, Habenberger, Kurtenbach, Dennenmoser et al., Identification of SRPK1 and SRPK2 as the major cellular protein kinases phosphorylating hepatitis B virus core protein, J Virol
De Lartigue, Polson, Feldman, Shokat, Tooze et al., PIKfyve regulation of endosome-linked pathways, Traffic
Dou, Li, Han, Zarlenga, Zhu et al., Cholesterol of lipid rafts is a key determinant for entry and post-entry control of porcine rotavirus infection, BMC Vet Res
Dowdle, Nyfeler, Nagel, Elling, Liu et al., Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo, Nat Cell Biol
Ericksen, Wu, Lu, Lehrer, Antibacterial activity and specificity of the six human {alpha}-defensins, Antimicrob Agents Chemother
Feng, Xu, Kovalev, Nagy, Recruitment of Vps34 PI3K and enrichment of PI3P phosphoinositide in the viral replication compartment is crucial for replication of a positive-strand RNA virus, PLoS Path
Ficarelli, Antzin-Anduetza, White, Firth, Sertkaya et al., CpG dinucleotides inhibit HIV-1 replication through zinc finger antiviral protein (ZAP)-dependent and -independent mechanisms, J Virol
Fonseca, Zakaria, Jia, Graber, Svitkin et al., La-related protein 1 (LARP1) represses terminal oligopyrimidine (TOP) mRNA translation downstream of mTOR complex 1 (mTORC1), J Biol Chem
Fu, He, Waheed, Dabbagh, Zhou et al., PSGL-1 restricts HIV-1 infectivity by blocking virus particle attachment to target cells, Proc Natl Acad Sci
Gaddy, Wong, Markowitz-Shulman, Poley, Regulation of the subcellular distribution of key cellular RNA-processing factors during permissive human cytomegalovirus infection, J Gen Virol
Gassen, Niemeyer, Muth, Corman, Martinelli et al., SKP2 attenuates autophagy through beclin1-ubiquitination and its inhibition reduces MERS-coronavirus infection, Nat Commun
He, Hetrick, Dabbagh, Akhrymuk, Kehn-Hall et al., PSGL-1 blocks SARS-CoV-2 S protein-mediated virus attachment and infection of target cells, bioRxiv
Heaton, Trimarco, Hamele, Harding, Tata et al., SRSF protein kinases 1 and 2 are essential host factors for human coronaviruses including SARS-CoV-2, bioRxiv
Hikmet, Méar, Edvinsson, Micke, Uhlén et al., The protein expression profile of ACE2 in human tissues, Mol Syst Biol
Hoffmann, Kleine-Weber, Pöhlmann, A Multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells, Mol Cell
Hoffmann, Kleine-Weber, Schroeder, Krüger, Herrler et al., SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor, Cell
Hoffmann, Sánchez-Rivera, Schneider, Luna, Soto-Feliciano et al., Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors, Cell Host Microbe
Huang, Bosch, Li, Li, Lee et al., SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells, J Biol Chem
Izaguirre, The proteolytic regulation of virus cell entry by furin and other proprotein convertases, Viruses
Jarvis, Allerston, Jia, Herzog, Garza-Garcia et al., Small molecule inhibitors of the neuropilin-1 vascular endothelial growth factor A (VEGF-A) interaction, J Med Chem
Jesus, Alexander, Anthony, Stacie, Christopher, Inhibitors of VPS34 and lipid metabolism suppress SARS-CoV-2 replication, bioRxiv
Johnson, Endapally, Vazquez, Infante, Radhakrishnan, Ostreolysin A and anthrolysin O use different mechanisms to control movement of cholesterol from the plasma membrane to the endoplasmic reticulum, J Biol Chem
Jones, Bevins, Paneth cells of the human small intestine express an antimicrobial peptide gene, J Biol Chem
Kang, Chou, Rothlauf, Liu, Soh et al., Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2, Proc Natl Acad Sci
Karakama, Sakamoto, Itsui, Nakagawa, Tasaka-Fujita et al., Inhibition of hepatitis C virus replication by a specific inhibitor of serine-arginine-rich protein kinase, Antimicrob Agents Chemother
Kawase, Shirato, Van Der Hoek, Taguchi, Matsuyama, Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry, J Virol
Kerns, Emerman, Malik, Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein, PLoS Genet
Klann, Bojkova, Tascher, Ciesek, Münch et al., Growth factor receptor signaling inhibition prevents SARS-CoV-2 replication, Mol Cell
Klein, Takahashi, Ma, Stagi, Zhou et al., Loss of TMEM106B ameliorates lysosomal and frontotemporal dementia-related phenotypes in progranulin-deficient mice, Neuron
Lang, Fellerer, Schwenk, Kuhn, Kremmer et al., Membrane orientation and subcellular localization of transmembrane protein 106B (TMEM106B), a major risk factor for frontotemporal lobar degeneration, J Biol Chem
Li, Deng, Wang, Ma, Aliyari et al., 25-Hydroxycholesterol protects host against zika virus infection and its associated microcephaly in a mouse model, Immunity
Li, Gu, Xu, Lysosomal ion channels as decoders of cellular signals, Trends Biochem Sci
Li, Hulswit, Widjaja, Raj, Mcbride et al., Identification of sialic acid-binding function for the middle east respiratory syndrome coronavirus spike glycoprotein, Proc Natl Acad Sci
Li, Li, Yamate, Li, Ikuta, Lipid rafts play an important role in the early stage of severe acute respiratory syndrome-coronavirus life cycle, Microbes Infect
Liefhebber, Hague, Zhang, Wakelam, Mclauchlan, Modulation of triglyceride and cholesterol ester synthesis impairs assembly of infectious hepatitis C virus, J Biol Chem
Liu, Aliyari, Chikere, Li, Marsden et al., Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol, Immunity
Liu, Fu, Wang, Li, Zhou et al., Proteomic profiling of HIV-1 infection of human CD4 T cells identifies PSGL-1 as an HIV restriction factor, Nat Microbiol
Lund, Kerr, Sakai, Li, Russell, cDNA cloning of mouse and human cholesterol 25-hydroxylases, polytopic membrane proteins that synthesize a potent oxysterol regulator of lipid metabolism, J Biol Chem
Luo, Yang, Song, Mechanisms and regulation of cholesterol homeostasis, Nat Rev Mol Cell Biol
Mar, Rinkenberger, Boys, Eitson, Mcdougal et al., LY6E mediates an evolutionarily conserved enhancement of virus infection by targeting a late entry step, Nat Commun
Meagher, Takata, Gonçalves-Carneiro, Keane, Rebendenne et al., Structure of the zinc-finger antiviral protein in complex with RNA reveals a mechanism for selective targeting of CG-rich viral sequences, Proc Natl Acad Sci
Milewska, Zarebski, Nowak, Stozek, Potempa et al., Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells, J Virol
Monteil, Kwon, Prado, Hagelkrüys, Wimmer et al., Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2, Cell
Nchioua, Kmiec, Müller, Conzelmann, Groß et al., SARS-CoV-2 is restricted by zinc finger antiviral protein despite preadaptation to the low-CpG environment in humans, mBio
Nicholson, Rademakers, What we know about TMEM106B in neurodegeneration, Acta Neuropathol
Nishimura, Shimojima, Tano, Miyamura, Wakita et al., Human P-selectin glycoprotein ligand-1 is a functional receptor for enterovirus 71, Nat Med
Niv, Defensin 5 for prevention of SARS-CoV-2 invasion and Covid-19 disease, Med Hypotheses
O'bryan, Frye, Cogswell, Neubauer, Kitch et al., Axl, a transforming gene isolated from primary human myeloid leukemia cells, encodes a novel receptor tyrosine kinase, Mol Cell Biol
Osuna-Ramos, Ruiz, Ángel, The role of host cholesterol during flavivirus infection, Front Cell Infect Microbiol
Park, Iwasaki, Type I and type III interferons-induction, signaling, evasion, and application to combat COVID-19, Cell Host Microbe
Peng, Lee, Tarn, Phosphorylation of the arginine/serine dipeptide-rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization, FEBS J
Pfaender, Mar, Michailidis, Kratzel, BoysV'kovski INP et al (2020) LY6E impairs coronavirus fusion and confers immune control of viral disease, Nat Microbiol
Prescott, Brimacombe, Hartley, Bell, Graham et al., Human papillomavirus type 1 E1^E4 protein is a potent inhibitor of the serine-arginine (SR) protein kinase SRPK1 and inhibits phosphorylation of host SR proteins and of the viral transcription and replication regulator E2, J Virol
Puelles, Lütgehetmann, Lindenmeyer, Sperhake, Wong et al., Multiorgan and renal tropism of SARS-CoV-2, N Engl J Med
Reggiori, Monastyrska, Verheije, Calì, Ulasli et al., Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication, Cell Host Microbe
Roderiquez, Oravecz, Yanagishita, Bou-Habib, Mostowski et al., Mediation of human immunodeficiency virus type 1 binding by interaction of cell surface heparan sulfate proteoglycans with the V3 region of envelope gp120-gp41, J Virol
Rutherford, Traer, Wassmer, Pattni, Bujny et al., The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport, J Cell Sci
Sako, Chang, Barone, Vachino, White et al., Expression cloning of a functional glycoprotein ligand for P-selectin, Cell
Sakurai, Kolokoltsov, Chen, Tidwell, Bauta et al., Ebola virus. Two-pore channels control ebola virus host cell entry and are drug targets for disease treatment, Science
Schmidt, Lareau, Keshishian, Ganskih, Schneider et al., The SARS-CoV-2 RNA-protein interactome in infected human cells, Nat Microbiol
Shang, Ye, Shi, Wan, Luo et al., Structural basis of receptor recognition by SARS-CoV-2, Nature
She, Zeng, Guo, Chen, Bai et al., Structural mechanisms of phospholipid activation of the human TPC2 channel, eLife
Shen, Azhar, Kraemer, SR-B1: a unique multifunctional receptor for cholesterol influx and efflux, Annu Rev Physiol
Shulla, Heald-Sargent, Subramanya, Zhao, Perlman et al., A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry, J Virol
Simmons, Gosalia, Rennekamp, Reeves, Diamond et al., Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry, Proc Natl Acad Sci
Simpson, Loh, Ullah, Lynch, Werder et al., Respiratory syncytial virus infection promotes necroptosis and HMGB1 release by airway epithelial cells, Am J Respir Crit Care Med
Snijder, Limpens, De Wilde, Jong, Zevenhoven-Dobbe et al., A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis, PLoS Biol
Somers, Tang, Shaw, Camphausen, Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P-and E-selectin bound to SLe(X) and PSGL-1, Cell
Su, Zhou, Chen, Wang, Qian et al., Cideb controls sterol-regulated ER export of SREBP/ SCAP by promoting cargo loading at ER exit sites, EMBO J
Sungnak, Huang, Bécavin, Berg, Queen et al., SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes, Nat Med
Takamatsu, Krähling, Kolesnikova, Halwe, Lier et al., Serine-arginine protein kinase 1 regulates ebola virus transcription, mBio
Takata, Gonçalves-Carneiro, Zang, Soll, York et al., CG dinucleotide suppression enables antiviral defence targeting non-self RNA, Nature
Teesalu, Sugahara, Kotamraju, Ruoslahti, C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration, Proc Natl Acad Sci
Tunnicliffe, Hu, Wu, Levy, Mould et al., Molecular mechanism of SR protein kinase 1 inhibition by the herpes virus protein ICP27, mBio
Walls, Park, Tortorici, Wall, Mcguire et al., Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein, Cell
Wang, Chen, Zhang, Deng, Lian et al., CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells, Signal Transduct Target Ther
Wang, Li, Hui, Tiwari, Zhang et al., Cholesterol 25-hydroxylase inhibits SARS-CoV-2 and other coronaviruses by depleting membrane cholesterol, EMBO J
Wang, Lin, Dyck, Yeakley, Songyang et al., SRPK2: a differentially expressed SR proteinspecific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells, J Cell Biol
Wang, Qiu, Hou, Deng, Xu et al., AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells, Cell Res
Wang, Shen, Gohain, Tolbert, Chen et al., Design of a potent antibiotic peptide based on the active region of human defensin 5, J Med Chem
Wang, Simoneau, Kulsuptrakul, Bouhaddou, Travisano et al., Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses, Cell
Wang, Wang, Li, Wei, Zhao et al., Human intestinal defensin 5 inhibits SARS-CoV-2 invasion by cloaking ACE2, Gastroenterology
Wang, Xu, Lin, Deng, Zhou et al., GPS 50: an update on the prediction of kinase-specific phosphorylation sites in proteins, Genomics Proteomics Bioinform
Wei, Wan, Yan, Wang, Zhang et al., HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry, Nat Metab
Woo, Huang, Lau, Yuen, Cytosine deamination and selection of CpG suppressed clones are the two major independent biological forces that shape codon usage bias in coronaviruses, Virology
Wrapp, Wang, Corbett, Goldsmith, Hsieh et al., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation, Science
Wudunn, Spear, Initial interaction of herpes simplex virus with cells is binding to heparan sulfate, J Virol
Xia, Extreme genomic CpG deficiency in SARS-CoV-2 and evasion of host antiviral defense, Mol Biol Evol
Xia, Lan, Su, Wang, Xu et al., The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin, Signal Transduct Target Ther
Yu, Liu, Emerging role of LY6E in virus-host interactions, Viruses
Yuen, Wong, Mak, Wang, Chu et al., Suppression of SARS-CoV-2 infection in ex-vivo human lung tissues by targeting class III phosphoinositide 3-kinase, J Med Virol
Yunqing, Jun, Xinyu, Hai, Yuyan et al., Interaction network of SARS-CoV-2 with host receptome through spike protein, bioRxiv
Zhao, Praissman, Grant, Cai, Xiao et al., Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor, Cell Host Microbe
Zhao, Zheng, Chen, Zheng, Li et al., LY6E restricts entry of human coronaviruses, including currently pandemic SARS-CoV-2, J Virol
Zhong, Ding, Adams, Ghosh, Fu, Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones, Genes Dev
Zhou, Xu, Castiglione, Soiberman, Eberhart et al., ACE2 and TMPRSS2 are expressed on the human ocular surface, suggesting susceptibility to SARS-CoV-2 infection, Ocul Surf
Zhu, Feng, Hu, Wang, Yu et al., A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry, Nat Commun
Ziegler, Allon, Nyquist, Mbano, Miao et al., SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues, Cell
DOI record:
{
"DOI": "10.1007/s11262-021-01869-2",
"ISSN": [
"0920-8569",
"1572-994X"
],
"URL": "http://dx.doi.org/10.1007/s11262-021-01869-2",
"alternative-id": [
"1869"
],
"assertion": [
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Received",
"name": "received",
"order": 1,
"value": "7 June 2021"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "28 August 2021"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "First Online",
"name": "first_online",
"order": 3,
"value": "11 September 2021"
},
{
"group": {
"label": "Declarations",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 1
},
{
"group": {
"label": "Conflict of interest",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 2,
"value": "The authors declare no conflict of interest."
},
{
"group": {
"label": "Informed consent",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 3,
"value": "Not applicable."
},
{
"group": {
"label": "Research involving human participants and/or animals",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 4,
"value": "Not applicable."
},
{
"label": "Free to read",
"name": "free",
"value": "This content has been made available to all."
}
],
"author": [
{
"affiliation": [],
"family": "Lv",
"given": "Lu",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0002-7015-9661",
"affiliation": [],
"authenticated-orcid": false,
"family": "Zhang",
"given": "Leiliang",
"sequence": "additional"
}
],
"container-title": "Virus Genes",
"container-title-short": "Virus Genes",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"link.springer.com"
]
},
"created": {
"date-parts": [
[
2021,
9,
12
]
],
"date-time": "2021-09-12T12:04:13Z",
"timestamp": 1631448253000
},
"deposited": {
"date-parts": [
[
2022,
10,
27
]
],
"date-time": "2022-10-27T19:29:11Z",
"timestamp": 1666898951000
},
"funder": [
{
"award": [
"2016YFD0500300"
],
"name": "National Key Plan for Research and Development of China"
},
{
"DOI": "10.13039/501100001809",
"award": [
"81871663",
"82072270"
],
"doi-asserted-by": "publisher",
"name": "National Natural Science Foundation of China"
},
{
"award": [
"2019LJ001"
],
"name": "Academic promotion programme of Shandong First Medical University"
}
],
"indexed": {
"date-parts": [
[
2024,
2,
3
]
],
"date-time": "2024-02-03T19:01:56Z",
"timestamp": 1706986916647
},
"is-referenced-by-count": 11,
"issue": "6",
"issued": {
"date-parts": [
[
2021,
9,
11
]
]
},
"journal-issue": {
"issue": "6",
"published-print": {
"date-parts": [
[
2021,
12
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://www.springer.com/tdm",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
9,
11
]
],
"date-time": "2021-09-11T00:00:00Z",
"timestamp": 1631318400000
}
},
{
"URL": "https://www.springer.com/tdm",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
9,
11
]
],
"date-time": "2021-09-11T00:00:00Z",
"timestamp": 1631318400000
}
}
],
"link": [
{
"URL": "https://link.springer.com/content/pdf/10.1007/s11262-021-01869-2.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://link.springer.com/article/10.1007/s11262-021-01869-2/fulltext.html",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://link.springer.com/content/pdf/10.1007/s11262-021-01869-2.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"page": "475-488",
"prefix": "10.1007",
"published": {
"date-parts": [
[
2021,
9,
11
]
]
},
"published-online": {
"date-parts": [
[
2021,
9,
11
]
]
},
"published-print": {
"date-parts": [
[
2021,
12
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference": [
{
"DOI": "10.1016/S0140-6736(20)30183-5",
"author": "C Huang",
"doi-asserted-by": "publisher",
"first-page": "497",
"issue": "10223",
"journal-title": "Lancet",
"key": "1869_CR1",
"unstructured": "Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y et al (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan China. Lancet 395(10223):497–506",
"volume": "395",
"year": "2020"
},
{
"DOI": "10.1093/jtm/taaa021",
"author": "Y Liu",
"doi-asserted-by": "publisher",
"first-page": "taaa021",
"issue": "2",
"journal-title": "J Travel Med",
"key": "1869_CR2",
"unstructured": "Liu Y, Gayle AA, Wilder-Smith A, Rocklöv J (2020) The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med 27(2):taaa021",
"volume": "27",
"year": "2020"
},
{
"DOI": "10.1016/j.chom.2020.02.001",
"author": "A Wu",
"doi-asserted-by": "publisher",
"first-page": "325",
"issue": "3",
"journal-title": "Cell Host Microbe",
"key": "1869_CR3",
"unstructured": "Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P et al (2020) Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27(3):325–328",
"volume": "27",
"year": "2020"
},
{
"DOI": "10.1016/S0140-6736(20)30251-8",
"author": "R Lu",
"doi-asserted-by": "publisher",
"first-page": "565",
"issue": "10224",
"journal-title": "Lancet",
"key": "1869_CR4",
"unstructured": "Lu R, Zhao X, Li J, Niu P, Yang B, Wu H et al (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395(10224):565–574",
"volume": "395",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2012-7",
"author": "P Zhou",
"doi-asserted-by": "publisher",
"first-page": "270",
"issue": "7798",
"journal-title": "Nature",
"key": "1869_CR5",
"unstructured": "Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W et al (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798):270–273",
"volume": "579",
"year": "2020"
},
{
"DOI": "10.3389/fimmu.2020.01022",
"author": "Y Liang",
"doi-asserted-by": "publisher",
"first-page": "1022",
"journal-title": "Front Immunol",
"key": "1869_CR6",
"unstructured": "Liang Y, Wang ML, Chien CS, Yarmishyn AA, Yang YP, Lai WY et al (2020) Highlight of immune pathogenic response and hematopathologic effect in SARS-CoV, MERS-CoV, and SARS-Cov-2 infection. Front Immunol 11:1022",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.03.045",
"author": "Q Wang",
"doi-asserted-by": "publisher",
"first-page": "894",
"issue": "4",
"journal-title": "Cell",
"key": "1869_CR7",
"unstructured": "Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z et al (2020) Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181(4):894-904.e9",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1038/s41564-020-0688-y",
"author": "M Letko",
"doi-asserted-by": "publisher",
"first-page": "562",
"issue": "4",
"journal-title": "Nat Microbiol",
"key": "1869_CR8",
"unstructured": "Letko M, Marzi A, Munster V (2020) Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5(4):562–569",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.1073/pnas.2003138117",
"author": "J Shang",
"doi-asserted-by": "publisher",
"first-page": "11727",
"issue": "21",
"journal-title": "Proc Natl Acad Sci USA",
"key": "1869_CR9",
"unstructured": "Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A et al (2020) Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA 117(21):11727–11734",
"volume": "117",
"year": "2020"
},
{
"DOI": "10.1038/s41467-020-15562-9",
"author": "X Ou",
"doi-asserted-by": "publisher",
"first-page": "1620",
"issue": "1",
"journal-title": "Nat Commun",
"key": "1869_CR10",
"unstructured": "Ou X, Liu Y, Lei X, Li P, Mi D, Ren L et al (2020) Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 11(1):1620",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2286-9",
"author": "DE Gordon",
"doi-asserted-by": "publisher",
"first-page": "459",
"issue": "7816",
"journal-title": "Nature",
"key": "1869_CR11",
"unstructured": "Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM et al (2020) A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583(7816):459–468",
"volume": "583",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.12.006",
"author": "WM Schneider",
"doi-asserted-by": "publisher",
"first-page": "120",
"issue": "1",
"journal-title": "Cell",
"key": "1869_CR12",
"unstructured": "Schneider WM, Luna JM, Hoffmann HH, Sánchez-Rivera FJ, Leal AA, Ashbrook AW et al (2021) Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell 184(1):120–32.e14",
"volume": "184",
"year": "2021"
},
{
"author": "J Wei",
"first-page": "31392",
"issue": "20",
"journal-title": "Cell",
"key": "1869_CR13",
"unstructured": "Wei J, Alfajaro MM, DeWeirdt PC, Hanna RE, Lu-Culligan WJ, Cai WL et al (2020) Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell S0092–8674(20):31392–31401",
"volume": "S0092–8674",
"year": "2020"
},
{
"author": "Z Daniloski",
"first-page": "31394",
"issue": "20",
"journal-title": "Cell",
"key": "1869_CR14",
"unstructured": "Daniloski Z, Jordan TX, Wessels HH, Hoagland DA, Kasela S, Legut M et al (2020) Identification of required host factors for SARS-CoV-2 infection in human cells. Cell S0092–8674(20):31394–31395",
"volume": "S0092–8674",
"year": "2020"
},
{
"DOI": "10.1038/nature02145",
"author": "W Li",
"doi-asserted-by": "publisher",
"first-page": "450",
"issue": "6965",
"journal-title": "Nature",
"key": "1869_CR15",
"unstructured": "Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA et al (2003) Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426(6965):450–454",
"volume": "426",
"year": "2003"
},
{
"DOI": "10.1073/pnas.0409465102",
"author": "H Hofmann",
"doi-asserted-by": "publisher",
"first-page": "7988",
"issue": "22",
"journal-title": "Proc Natl Acad Sci USA",
"key": "1869_CR16",
"unstructured": "Hofmann H, Pyrc K, van der Hoek L, Geier M, Berkhout B, Pöhlmann S (2005) Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci USA 102(22):7988–7993",
"volume": "102",
"year": "2005"
},
{
"DOI": "10.1126/science.abb2507",
"author": "D Wrapp",
"doi-asserted-by": "publisher",
"first-page": "1260",
"issue": "6483",
"journal-title": "Science",
"key": "1869_CR17",
"unstructured": "Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O et al (2020) Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367(6483):1260–1263",
"volume": "367",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2179-y",
"author": "J Shang",
"doi-asserted-by": "publisher",
"first-page": "221",
"issue": "7807",
"journal-title": "Nature",
"key": "1869_CR18",
"unstructured": "Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H et al (2020) Structural basis of receptor recognition by SARS-CoV-2. Nature 581(7807):221–224",
"volume": "581",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.02.052",
"author": "M Hoffmann",
"doi-asserted-by": "publisher",
"first-page": "271",
"issue": "2",
"journal-title": "Cell",
"key": "1869_CR19",
"unstructured": "Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S et al (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181(2):271–80.e8",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.04.004",
"author": "V Monteil",
"doi-asserted-by": "publisher",
"first-page": "905",
"issue": "4",
"journal-title": "Cell",
"key": "1869_CR20",
"unstructured": "Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, Stahl M et al (2020) Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell 181(4):905–13.e7",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.15252/msb.20209610",
"author": "F Hikmet",
"doi-asserted-by": "publisher",
"first-page": "e9610",
"issue": "7",
"journal-title": "Mol Syst Biol",
"key": "1869_CR21",
"unstructured": "Hikmet F, Méar L, Edvinsson Å, Micke P, Uhlén M, Lindskog C (2020) The protein expression profile of ACE2 in human tissues. Mol Syst Biol 16(7):e9610",
"volume": "16",
"year": "2020"
},
{
"DOI": "10.1038/nature10606",
"author": "C Crosnier",
"doi-asserted-by": "publisher",
"first-page": "534",
"issue": "7378",
"journal-title": "Nature",
"key": "1869_CR22",
"unstructured": "Crosnier C, Bustamante LY, Bartholdson SJ, Bei AK, Theron M, Uchikawa M et al (2011) Basigin is a receptor essential for erythrocyte invasion by plasmodium falciparum. Nature 480(7378):534–537",
"volume": "480",
"year": "2011"
},
{
"DOI": "10.1086/427811",
"author": "Z Chen",
"doi-asserted-by": "publisher",
"first-page": "755",
"issue": "5",
"journal-title": "J Infect Dis",
"key": "1869_CR23",
"unstructured": "Chen Z, Mi L, Xu J, Yu J, Wang X, Jiang J et al (2005) Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. J Infect Dis 191(5):755–760",
"volume": "191",
"year": "2005"
},
{
"DOI": "10.1038/s41392-020-00426-x",
"author": "K Wang",
"doi-asserted-by": "publisher",
"first-page": "283",
"issue": "1",
"journal-title": "Signal Transduct Target Ther",
"key": "1869_CR24",
"unstructured": "Wang K, Chen W, Zhang Z, Deng Y, Lian JQ, Du P et al (2020) CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Target Ther 5(1):283",
"volume": "5",
"year": "2020"
},
{
"author": "JP O'Bryan",
"first-page": "5016",
"issue": "10",
"journal-title": "Mol Cell Biol",
"key": "1869_CR25",
"unstructured": "O’Bryan JP, Frye RA, Cogswell PC, Neubauer A, Kitch B, Prokop C et al (1991) Axl, a transforming gene isolated from primary human myeloid leukemia cells, encodes a novel receptor tyrosine kinase. Mol Cell Biol 11(10):5016–5031",
"volume": "11",
"year": "1991"
},
{
"DOI": "10.1038/s41422-020-00460-y",
"author": "S Wang",
"doi-asserted-by": "publisher",
"first-page": "126",
"issue": "2",
"journal-title": "Cell Res",
"key": "1869_CR26",
"unstructured": "Wang S, Qiu Z, Hou Y, Deng X, Xu W, Zheng T et al (2021) AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res 31(2):126–140",
"volume": "31",
"year": "2021"
},
{
"DOI": "10.3390/v11070596",
"author": "V Cagno",
"doi-asserted-by": "publisher",
"first-page": "596",
"issue": "7",
"journal-title": "Viruses",
"key": "1869_CR27",
"unstructured": "Cagno V, Tseligka ED, Jones ST, Tapparel C (2019) Heparan sulfate proteoglycans and viral attachment: true receptors or adaptation bias? Viruses 11(7):596",
"volume": "11",
"year": "2019"
},
{
"DOI": "10.1128/jvi.63.1.52-58.1989",
"author": "D WuDunn",
"doi-asserted-by": "publisher",
"first-page": "52",
"issue": "1",
"journal-title": "J Virol",
"key": "1869_CR28",
"unstructured": "WuDunn D, Spear PG (1989) Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol 63(1):52–58",
"volume": "63",
"year": "1989"
},
{
"DOI": "10.1128/jvi.69.4.2233-2239.1995",
"author": "G Roderiquez",
"doi-asserted-by": "publisher",
"first-page": "2233",
"issue": "4",
"journal-title": "J Virol",
"key": "1869_CR29",
"unstructured": "Roderiquez G, Oravecz T, Yanagishita M, Bou-Habib DC, Mostowski H, Norcross MA (1995) Mediation of human immunodeficiency virus type 1 binding by interaction of cell surface heparan sulfate proteoglycans with the V3 region of envelope gp120-gp41. J Virol 69(4):2233–2239",
"volume": "69",
"year": "1995"
},
{
"DOI": "10.1128/JVI.02078-14",
"author": "A Milewska",
"doi-asserted-by": "publisher",
"first-page": "13221",
"issue": "22",
"journal-title": "J Virol",
"key": "1869_CR30",
"unstructured": "Milewska A, Zarebski M, Nowak P, Stozek K, Potempa J, Pyrc K (2014) Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J Virol 88(22):13221–13230",
"volume": "88",
"year": "2014"
},
{
"DOI": "10.1016/j.cell.2020.09.033",
"author": "TM Clausen",
"doi-asserted-by": "publisher",
"first-page": "1043",
"issue": "4",
"journal-title": "Cell",
"key": "1869_CR31",
"unstructured": "Clausen TM, Sandoval DR, Spliid CB, Pihl J, Perrett HR, Painter CD et al (2020) SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell 183(4):1043–57.e15",
"volume": "183",
"year": "2020"
},
{
"DOI": "10.1038/s41467-020-20457-w",
"author": "H Chu",
"doi-asserted-by": "publisher",
"first-page": "134",
"issue": "1",
"journal-title": "Nat Commun",
"key": "1869_CR32",
"unstructured": "Chu H, Hu B, Huang X, Chai Y, Zhou D, Wang Y et al (2021) Host and viral determinants for efficient SARS-CoV-2 infection of the human lung. Nat Commun 12(1):134",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.02.058",
"author": "AC Walls",
"doi-asserted-by": "publisher",
"first-page": "281",
"issue": "2",
"journal-title": "Cell",
"key": "1869_CR33",
"unstructured": "Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D (2020) Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181(2):281–92.e6",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1016/j.molcel.2020.04.022",
"author": "M Hoffmann",
"doi-asserted-by": "publisher",
"first-page": "779",
"issue": "4",
"journal-title": "Mol Cell",
"key": "1869_CR34",
"unstructured": "Hoffmann M, Kleine-Weber H, Pöhlmann S (2020) A Multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell 78(4):779–84.e5",
"volume": "78",
"year": "2020"
},
{
"DOI": "10.1073/pnas.0908201106",
"author": "T Teesalu",
"doi-asserted-by": "publisher",
"first-page": "16157",
"issue": "38",
"journal-title": "Proc Natl Acad Sci USA",
"key": "1869_CR35",
"unstructured": "Teesalu T, Sugahara KN, Kotamraju VR, Ruoslahti E (2009) C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc Natl Acad Sci USA 106(38):16157–16162",
"volume": "106",
"year": "2009"
},
{
"DOI": "10.1126/science.abd3072",
"author": "JL Daly",
"doi-asserted-by": "publisher",
"first-page": "861",
"issue": "6518",
"journal-title": "Science",
"key": "1869_CR36",
"unstructured": "Daly JL, Simonetti B, Klein K, Chen KE, Williamson MK, Antón-Plágaro C et al (2020) Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 370(6518):861–865",
"volume": "370",
"year": "2020"
},
{
"DOI": "10.1021/jm901755g",
"author": "A Jarvis",
"doi-asserted-by": "publisher",
"first-page": "2215",
"issue": "5",
"journal-title": "J Med Chem",
"key": "1869_CR37",
"unstructured": "Jarvis A, Allerston CK, Jia H, Herzog B, Garza-Garcia A, Winfield N et al (2010) Small molecule inhibitors of the neuropilin-1 vascular endothelial growth factor A (VEGF-A) interaction. J Med Chem 53(5):2215–2226",
"volume": "53",
"year": "2010"
},
{
"DOI": "10.1126/science.abd2985",
"author": "L Cantuti-Castelvetri",
"doi-asserted-by": "publisher",
"first-page": "856",
"issue": "6518",
"journal-title": "Science",
"key": "1869_CR38",
"unstructured": "Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S et al (2020) Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370(6518):856–860",
"volume": "370",
"year": "2020"
},
{
"DOI": "10.1126/science.271.5248.518",
"author": "S Acton",
"doi-asserted-by": "publisher",
"first-page": "518",
"issue": "5248",
"journal-title": "Science",
"key": "1869_CR39",
"unstructured": "Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M (1996) Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science 271(5248):518–520",
"volume": "271",
"year": "1996"
},
{
"DOI": "10.1146/annurev-physiol-021317-121550",
"author": "WJ Shen",
"doi-asserted-by": "publisher",
"first-page": "95",
"journal-title": "Annu Rev Physiol",
"key": "1869_CR40",
"unstructured": "Shen WJ, Azhar S, Kraemer FB (2018) SR-B1: a unique multifunctional receptor for cholesterol influx and efflux. Annu Rev Physiol 80:95–116",
"volume": "80",
"year": "2018"
},
{
"DOI": "10.1016/j.virol.2017.07.024",
"author": "S Bajimaya",
"doi-asserted-by": "publisher",
"first-page": "234",
"journal-title": "Virology",
"key": "1869_CR41",
"unstructured": "Bajimaya S, Frankl T, Hayashi T, Takimoto T (2017) Cholesterol is required for stability and infectivity of influenza A and respiratory syncytial viruses. Virology 510:234–241",
"volume": "510",
"year": "2017"
},
{
"DOI": "10.3389/fcimb.2018.00388",
"author": "JF Osuna-Ramos",
"doi-asserted-by": "publisher",
"first-page": "388",
"journal-title": "Front Cell Infect Microbiol",
"key": "1869_CR42",
"unstructured": "Osuna-Ramos JF, Reyes-Ruiz JM, Del Ángel RM (2018) The role of host cholesterol during flavivirus infection. Front Cell Infect Microbiol 8:388",
"volume": "8",
"year": "2018"
},
{
"DOI": "10.1186/s12917-018-1366-7",
"author": "X Dou",
"doi-asserted-by": "publisher",
"first-page": "45",
"issue": "1",
"journal-title": "BMC Vet Res",
"key": "1869_CR43",
"unstructured": "Dou X, Li Y, Han J, Zarlenga DS, Zhu W, Ren X et al (2018) Cholesterol of lipid rafts is a key determinant for entry and post-entry control of porcine rotavirus infection. BMC Vet Res 14(1):45",
"volume": "14",
"year": "2018"
},
{
"DOI": "10.1016/j.micinf.2006.10.015",
"author": "GM Li",
"doi-asserted-by": "publisher",
"first-page": "96",
"issue": "1",
"journal-title": "Microbes Infect",
"key": "1869_CR44",
"unstructured": "Li GM, Li YG, Yamate M, Li SM, Ikuta K (2007) Lipid rafts play an important role in the early stage of severe acute respiratory syndrome-coronavirus life cycle. Microbes Infect 9(1):96–102",
"volume": "9",
"year": "2007"
},
{
"DOI": "10.1128/JVI.02199-08",
"author": "MT Catanese",
"doi-asserted-by": "publisher",
"first-page": "34",
"issue": "1",
"journal-title": "J Virol",
"key": "1869_CR45",
"unstructured": "Catanese MT, Ansuini H, Graziani R, Huby T, Moreau M, Ball JK et al (2010) Role of scavenger receptor class B type I in hepatitis C virus entry: kinetics and molecular determinants. J Virol 84(1):34–43",
"volume": "84",
"year": "2010"
},
{
"DOI": "10.1038/s42255-020-00324-0",
"author": "C Wei",
"doi-asserted-by": "publisher",
"first-page": "1391",
"issue": "12",
"journal-title": "Nat Metab",
"key": "1869_CR46",
"unstructured": "Wei C, Wan L, Yan Q, Wang X, Zhang J, Yang X et al (2020) HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat Metab 2(12):1391–1400",
"volume": "2",
"year": "2020"
},
{
"DOI": "10.1101/2020.09.09.287508",
"doi-asserted-by": "crossref",
"key": "1869_CR47",
"unstructured": "Gu Yunqing, Cao Jun, Zhang Xinyu, Gao Hai, Wang Yuyan, Wang Jia, et al (2020) Interaction network of SARS-CoV-2 with host receptome through spike protein. bioRxiv:2020.09.09.287508"
},
{
"DOI": "10.1056/NEJMc2011400",
"author": "VG Puelles",
"doi-asserted-by": "publisher",
"first-page": "590",
"issue": "6",
"journal-title": "N Engl J Med",
"key": "1869_CR48",
"unstructured": "Puelles VG, Lütgehetmann M, Lindenmeyer MT, Sperhake JP, Wong MN, Allweiss L et al (2020) Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med 383(6):590–592",
"volume": "383",
"year": "2020"
},
{
"DOI": "10.1038/s41587-020-0602-4",
"author": "RL Chua",
"doi-asserted-by": "publisher",
"first-page": "970",
"issue": "8",
"journal-title": "Nat Biotechnol",
"key": "1869_CR49",
"unstructured": "Chua RL, Lukassen S, Trump S, Hennig BP, Wendisch D, Pott F et al (2020) COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol 38(8):970–979",
"volume": "38",
"year": "2020"
},
{
"DOI": "10.1038/s41591-020-0868-6",
"author": "W Sungnak",
"doi-asserted-by": "publisher",
"first-page": "681",
"issue": "5",
"journal-title": "Nat Med",
"key": "1869_CR50",
"unstructured": "Sungnak W, Huang N, Bécavin C, Berg M, Queen R, Litvinukova M et al (2020) SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med 26(5):681–687",
"volume": "26",
"year": "2020"
},
{
"DOI": "10.1016/j.smim.2018.02.011",
"author": "U Andersson",
"doi-asserted-by": "publisher",
"first-page": "40",
"journal-title": "Semin Immunol",
"key": "1869_CR51",
"unstructured": "Andersson U, Yang H, Harris H (2018) High-mobility group box 1 protein (HMGB1) operates as an alarmin outside as well as inside cells. Semin Immunol 38:40–48",
"volume": "38",
"year": "2018"
},
{
"DOI": "10.1186/s10020-020-00172-4",
"author": "U Andersson",
"doi-asserted-by": "publisher",
"first-page": "42",
"issue": "1",
"journal-title": "Mol Med",
"key": "1869_CR52",
"unstructured": "Andersson U, Ottestad W, Tracey KJ (2020) Extracellular HMGB1: a therapeutic target in severe pulmonary inflammation including COVID-19? Mol Med 26(1):42",
"volume": "26",
"year": "2020"
},
{
"DOI": "10.1164/rccm.201906-1149OC",
"author": "J Simpson",
"doi-asserted-by": "publisher",
"first-page": "1358",
"issue": "11",
"journal-title": "Am J Respir Crit Care Med",
"key": "1869_CR53",
"unstructured": "Simpson J, Loh Z, Ullah MA, Lynch JP, Werder RB, Collinson N et al (2020) Respiratory syncytial virus infection promotes necroptosis and HMGB1 release by airway epithelial cells. Am J Respir Crit Care Med 201(11):1358–1371",
"volume": "201",
"year": "2020"
},
{
"DOI": "10.1038/s41467-021-21213-4",
"author": "Y Zhu",
"doi-asserted-by": "publisher",
"first-page": "961",
"issue": "1",
"journal-title": "Nat Commun",
"key": "1869_CR54",
"unstructured": "Zhu Y, Feng F, Hu G, Wang Y, Yu Y, Zhu Y et al (2021) A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat Commun 12(1):961",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1016/j.chom.2020.12.009",
"author": "HH Hoffmann",
"doi-asserted-by": "publisher",
"first-page": "267",
"issue": "2",
"journal-title": "Cell Host Microbe",
"key": "1869_CR55",
"unstructured": "Hoffmann HH, Sánchez-Rivera FJ, Schneider WM, Luna JM, Soto-Feliciano YM, Ashbrook AW et al (2021) Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe 29(2):267–80.e5",
"volume": "29",
"year": "2021"
},
{
"DOI": "10.3389/fimmu.2020.552925",
"author": "NM Beeraka",
"doi-asserted-by": "publisher",
"first-page": "552925",
"journal-title": "Front Immunol",
"key": "1869_CR56",
"unstructured": "Beeraka NM, Sadhu SP, Madhunapantula SV, Rao Pragada R, Svistunov AA, Nikolenko VN et al (2020) Strategies for targeting SARS CoV-2: small molecule inhibitors-the current status. Front Immunol 11:552925",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1128/JVI.02062-10",
"author": "A Shulla",
"doi-asserted-by": "publisher",
"first-page": "873",
"issue": "2",
"journal-title": "J Virol",
"key": "1869_CR57",
"unstructured": "Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T (2011) A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol 85(2):873–882",
"volume": "85",
"year": "2011"
},
{
"DOI": "10.1128/JVI.00094-12",
"author": "M Kawase",
"doi-asserted-by": "publisher",
"first-page": "6537",
"issue": "12",
"journal-title": "J Virol",
"key": "1869_CR58",
"unstructured": "Kawase M, Shirato K, van der Hoek L, Taguchi F, Matsuyama S (2012) Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J Virol 86(12):6537–6545",
"volume": "86",
"year": "2012"
},
{
"DOI": "10.1016/j.cell.2020.04.035",
"author": "CGK Ziegler",
"doi-asserted-by": "publisher",
"first-page": "1016",
"issue": "5",
"journal-title": "Cell",
"key": "1869_CR59",
"unstructured": "Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN et al (2020) SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 181(5):1016–35.e19",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1016/j.jtos.2020.06.007",
"author": "L Zhou",
"doi-asserted-by": "publisher",
"first-page": "537",
"issue": "4",
"journal-title": "Ocul Surf",
"key": "1869_CR60",
"unstructured": "Zhou L, Xu Z, Castiglione GM, Soiberman US, Eberhart CG, Duh EJ (2020) ACE2 and TMPRSS2 are expressed on the human ocular surface, suggesting susceptibility to SARS-CoV-2 infection. Ocul Surf 18(4):537–544",
"volume": "18",
"year": "2020"
},
{
"DOI": "10.1002/cti2.1073",
"author": "E Braun",
"doi-asserted-by": "publisher",
"first-page": "e1073",
"issue": "8",
"journal-title": "Clin Transl Immunol",
"key": "1869_CR61",
"unstructured": "Braun E, Sauter D (2019) Furin-mediated protein processing in infectious diseases and cancer. Clin Transl Immunol 8(8):e1073",
"volume": "8",
"year": "2019"
},
{
"DOI": "10.3390/v11090837",
"author": "G Izaguirre",
"doi-asserted-by": "publisher",
"first-page": "837",
"issue": "9",
"journal-title": "Viruses",
"key": "1869_CR62",
"unstructured": "Izaguirre G (2019) The proteolytic regulation of virus cell entry by furin and other proprotein convertases. Viruses 11(9):837",
"volume": "11",
"year": "2019"
},
{
"DOI": "10.1016/j.celrep.2020.108254",
"author": "YW Cheng",
"doi-asserted-by": "publisher",
"first-page": "1254",
"issue": "2",
"journal-title": "Cell Rep",
"key": "1869_CR63",
"unstructured": "Cheng YW, Chao TL, Li CL, Chiu MF, Kao HC, Wang SH et al (2020) Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects. Cell Rep 33(2):1254",
"volume": "33",
"year": "2020"
},
{
"DOI": "10.1038/s41392-020-0184-0",
"author": "S Xia",
"doi-asserted-by": "publisher",
"first-page": "92",
"issue": "1",
"journal-title": "Signal Transduct Target Ther",
"key": "1869_CR64",
"unstructured": "Xia S, Lan Q, Su S, Wang X, Xu W, Liu Z et al (2020) The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct Target Ther 5(1):92",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.1073/pnas.0505577102",
"author": "G Simmons",
"doi-asserted-by": "publisher",
"first-page": "11876",
"issue": "33",
"journal-title": "Proc Natl Acad Sci USA",
"key": "1869_CR65",
"unstructured": "Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P (2005) Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA 102(33):11876–11881",
"volume": "102",
"year": "2005"
},
{
"DOI": "10.1074/jbc.M508381200",
"author": "IC Huang",
"doi-asserted-by": "publisher",
"first-page": "3198",
"issue": "6",
"journal-title": "J Biol Chem",
"key": "1869_CR66",
"unstructured": "Huang IC, Bosch BJ, Li F, Li W, Lee KH, Ghiran S et al (2006) SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J Biol Chem 281(6):3198–3203",
"volume": "281",
"year": "2006"
},
{
"DOI": "10.1111/j.1600-0854.2009.00915.x",
"author": "J de Lartigue",
"doi-asserted-by": "publisher",
"first-page": "883",
"issue": "7",
"journal-title": "Traffic",
"key": "1869_CR67",
"unstructured": "de Lartigue J, Polson H, Feldman M, Shokat K, Tooze SA, Urbé S et al (2009) PIKfyve regulation of endosome-linked pathways. Traffic 10(7):883–893",
"volume": "10",
"year": "2009"
},
{
"DOI": "10.1242/jcs.03153",
"author": "AC Rutherford",
"doi-asserted-by": "publisher",
"first-page": "3944",
"journal-title": "J Cell Sci",
"key": "1869_CR68",
"unstructured": "Rutherford AC, Traer C, Wassmer T, Pattni K, Bujny MV, Carlton JG et al (2006) The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J Cell Sci 119:3944–3957",
"volume": "119",
"year": "2006"
},
{
"DOI": "10.1073/pnas.2007837117",
"author": "YL Kang",
"doi-asserted-by": "publisher",
"first-page": "20803",
"issue": "34",
"journal-title": "Proc Natl Acad Sci USA",
"key": "1869_CR69",
"unstructured": "Kang YL, Chou YY, Rothlauf PW, Liu Z, Soh TK, Cureton D et al (2020) Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. Proc Natl Acad Sci USA 117(34):20803–20813",
"volume": "117",
"year": "2020"
},
{
"DOI": "10.7554/eLife.45222",
"author": "J She",
"doi-asserted-by": "publisher",
"first-page": "e45222",
"journal-title": "eLife",
"key": "1869_CR70",
"unstructured": "She J, Zeng W, Guo J, Chen Q, Bai XC, Jiang Y (2019) Structural mechanisms of phospholipid activation of the human TPC2 channel. eLife 8:e45222",
"volume": "8",
"year": "2019"
},
{
"DOI": "10.1016/j.tibs.2018.10.006",
"author": "P Li",
"doi-asserted-by": "publisher",
"first-page": "110",
"issue": "2",
"journal-title": "Trends Biochem Sci",
"key": "1869_CR71",
"unstructured": "Li P, Gu M, Xu H (2019) Lysosomal ion channels as decoders of cellular signals. Trends Biochem Sci 44(2):110–124",
"volume": "44",
"year": "2019"
},
{
"DOI": "10.1126/science.1258758",
"author": "Y Sakurai",
"doi-asserted-by": "publisher",
"first-page": "995",
"issue": "6225",
"journal-title": "Science",
"key": "1869_CR72",
"unstructured": "Sakurai Y, Kolokoltsov AA, Chen CC, Tidwell MW, Bauta WE, Klugbauer N et al (2015) Ebola virus. Two-pore channels control ebola virus host cell entry and are drug targets for disease treatment. Science 347(6225):995–8",
"volume": "347",
"year": "2015"
},
{
"DOI": "10.1074/jbc.M112.365098",
"author": "CM Lang",
"doi-asserted-by": "publisher",
"first-page": "19355",
"issue": "23",
"journal-title": "J Biol Chem",
"key": "1869_CR73",
"unstructured": "Lang CM, Fellerer K, Schwenk BM, Kuhn PH, Kremmer E, Edbauer D et al (2012) Membrane orientation and subcellular localization of transmembrane protein 106B (TMEM106B), a major risk factor for frontotemporal lobar degeneration. J Biol Chem 287(23):19355–19365",
"volume": "287",
"year": "2012"
},
{
"DOI": "10.1093/hmg/dds475",
"author": "OA Brady",
"doi-asserted-by": "publisher",
"first-page": "685",
"issue": "4",
"journal-title": "Hum Mol Genet",
"key": "1869_CR74",
"unstructured": "Brady OA, Zheng Y, Murphy K, Huang M, Hu F (2013) The frontotemporal lobar degeneration risk factor, TMEM106B, regulates lysosomal morphology and function. Hum Mol Genet 22(4):685–695",
"volume": "22",
"year": "2013"
},
{
"DOI": "10.1007/s00401-016-1610-9",
"author": "AM Nicholson",
"doi-asserted-by": "publisher",
"first-page": "639",
"issue": "5",
"journal-title": "Acta Neuropathol",
"key": "1869_CR75",
"unstructured": "Nicholson AM, Rademakers R (2016) What we know about TMEM106B in neurodegeneration. Acta Neuropathol 132(5):639–651",
"volume": "132",
"year": "2016"
},
{
"DOI": "10.1016/j.cell.2020.12.004",
"author": "R Wang",
"doi-asserted-by": "publisher",
"first-page": "106",
"issue": "1",
"journal-title": "Cell",
"key": "1869_CR76",
"unstructured": "Wang R, Simoneau CR, Kulsuptrakul J, Bouhaddou M, Travisano KA, Hayashi JM et al (2021) Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell 184(1):106–19.e14",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1038/s41588-021-00805-2",
"author": "J Baggen",
"doi-asserted-by": "publisher",
"first-page": "435",
"journal-title": "Nat Genet",
"key": "1869_CR77",
"unstructured": "Baggen J, Persoons L, Vanstreels E, Jansen S, Van Looveren D, Boeckx B et al (2021) Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat Genet 53:435–444",
"volume": "53",
"year": "2021"
},
{
"DOI": "10.1038/nature10348",
"author": "JE Carette",
"doi-asserted-by": "publisher",
"first-page": "340",
"issue": "7364",
"journal-title": "Nature",
"key": "1869_CR78",
"unstructured": "Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N et al (2011) Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477(7364):340–343",
"volume": "477",
"year": "2011"
},
{
"DOI": "10.1016/j.neuron.2017.06.026",
"author": "ZA Klein",
"doi-asserted-by": "publisher",
"first-page": "281",
"issue": "2",
"journal-title": "Neuron",
"key": "1869_CR79",
"unstructured": "Klein ZA, Takahashi H, Ma M, Stagi M, Zhou M, Lam TT et al (2017) Loss of TMEM106B ameliorates lysosomal and frontotemporal dementia-related phenotypes in progranulin-deficient mice. Neuron 95(2):281–96.e6",
"volume": "95",
"year": "2017"
},
{
"DOI": "10.1021/bi4010864",
"author": "BE Aubol",
"doi-asserted-by": "publisher",
"first-page": "7595",
"issue": "43",
"journal-title": "Biochemistry",
"key": "1869_CR80",
"unstructured": "Aubol BE, Jamros MA, McGlone ML, Adams JA (2013) Splicing kinase SRPK1 conforms to the landscape of its SR protein substrate. Biochemistry 52(43):7595–7605",
"volume": "52",
"year": "2013"
},
{
"DOI": "10.1101/gad.1752109",
"author": "XY Zhong",
"doi-asserted-by": "publisher",
"first-page": "482",
"issue": "4",
"journal-title": "Genes Dev",
"key": "1869_CR81",
"unstructured": "Zhong XY, Ding JH, Adams JA, Ghosh G, Fu XD (2009) Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev 23(4):482–495",
"volume": "23",
"year": "2009"
},
{
"DOI": "10.1083/jcb.140.4.737",
"author": "HY Wang",
"doi-asserted-by": "publisher",
"first-page": "737",
"issue": "4",
"journal-title": "J Cell Biol",
"key": "1869_CR82",
"unstructured": "Wang HY, Lin W, Dyck JA, Yeakley JM, Songyang Z, Cantley LC et al (1998) SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J Cell Biol 140(4):737–750",
"volume": "140",
"year": "1998"
},
{
"DOI": "10.1128/mBio.02565-19",
"author": "Y Takamatsu",
"doi-asserted-by": "publisher",
"first-page": "e02565",
"issue": "1",
"journal-title": "mBio",
"key": "1869_CR83",
"unstructured": "Takamatsu Y, Krähling V, Kolesnikova L, Halwe S, Lier C, Baumeister S et al (2020) Serine-arginine protein kinase 1 regulates ebola virus transcription. mBio 11(1):e02565-19",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1128/AAC.00113-10",
"author": "Y Karakama",
"doi-asserted-by": "publisher",
"first-page": "3179",
"issue": "8",
"journal-title": "Antimicrob Agents Chemother",
"key": "1869_CR84",
"unstructured": "Karakama Y, Sakamoto N, Itsui Y, Nakagawa M, Tasaka-Fujita M, Nishimura-Sakurai Y et al (2010) Inhibition of hepatitis C virus replication by a specific inhibitor of serine-arginine-rich protein kinase. Antimicrob Agents Chemother 54(8):3179–3186",
"volume": "54",
"year": "2010"
},
{
"DOI": "10.1099/vir.0.020313-0",
"author": "CE Gaddy",
"doi-asserted-by": "publisher",
"first-page": "1547",
"journal-title": "J Gen Virol",
"key": "1869_CR85",
"unstructured": "Gaddy CE, Wong DS, Markowitz-Shulman A, Colberg-Poley AM (2010) Regulation of the subcellular distribution of key cellular RNA-processing factors during permissive human cytomegalovirus infection. J Gen Virol 91:1547–1559",
"volume": "91",
"year": "2010"
},
{
"DOI": "10.1128/JVI.02029-14",
"author": "EL Prescott",
"doi-asserted-by": "publisher",
"first-page": "12599",
"issue": "21",
"journal-title": "J Virol",
"key": "1869_CR86",
"unstructured": "Prescott EL, Brimacombe CL, Hartley M, Bell I, Graham S, Roberts S (2014) Human papillomavirus type 1 E1^E4 protein is a potent inhibitor of the serine-arginine (SR) protein kinase SRPK1 and inhibits phosphorylation of host SR proteins and of the viral transcription and replication regulator E2. J Virol 88(21):12599–12611",
"volume": "88",
"year": "2014"
},
{
"DOI": "10.1128/mBio.02551-19",
"author": "RB Tunnicliffe",
"doi-asserted-by": "publisher",
"first-page": "e02551",
"issue": "5",
"journal-title": "mBio",
"key": "1869_CR87",
"unstructured": "Tunnicliffe RB, Hu WK, Wu MY, Levy C, Mould AP, McKenzie EA et al (2019) Molecular mechanism of SR protein kinase 1 inhibition by the herpes virus protein ICP27. mBio 10(5):e02551-19",
"volume": "10",
"year": "2019"
},
{
"DOI": "10.1128/JVI.76.16.8124-8137.2002",
"author": "H Daub",
"doi-asserted-by": "publisher",
"first-page": "8124",
"issue": "16",
"journal-title": "J Virol",
"key": "1869_CR88",
"unstructured": "Daub H, Blencke S, Habenberger P, Kurtenbach A, Dennenmoser J, Wissing J et al (2002) Identification of SRPK1 and SRPK2 as the major cellular protein kinases phosphorylating hepatitis B virus core protein. J Virol 76(16):8124–8137",
"volume": "76",
"year": "2002"
},
{
"DOI": "10.1111/j.1742-4658.2008.06564.x",
"author": "TY Peng",
"doi-asserted-by": "publisher",
"first-page": "4152",
"issue": "16",
"journal-title": "FEBS J",
"key": "1869_CR89",
"unstructured": "Peng TY, Lee KR, Tarn WY (2008) Phosphorylation of the arginine/serine dipeptide-rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. FEBS J 275(16):4152–4163",
"volume": "275",
"year": "2008"
},
{
"key": "1869_CR90",
"unstructured": "Heaton BE, Trimarco JD, Hamele CE, Harding AT, Tata A, Zhu X, et al (2020) SRSF protein kinases 1 and 2 are essential host factors for human coronaviruses including SARS-CoV-2. bioRxiv:2020.08.14.251207"
},
{
"DOI": "10.1016/j.gpb.2020.01.001",
"author": "C Wang",
"doi-asserted-by": "publisher",
"first-page": "72",
"issue": "1",
"journal-title": "Genomics Proteomics Bioinform",
"key": "1869_CR91",
"unstructured": "Wang C, Xu H, Lin S, Deng W, Zhou J, Zhang Y et al (2020) GPS 50: an update on the prediction of kinase-specific phosphorylation sites in proteins. Genomics Proteomics Bioinform 18(1):72–80",
"volume": "18",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.06.034",
"author": "M Bouhaddou",
"doi-asserted-by": "publisher",
"first-page": "685",
"issue": "3",
"journal-title": "Cell",
"key": "1869_CR92",
"unstructured": "Bouhaddou M, Memon D, Meyer B, White KM, Rezelj VV, Correa Marrero M et al (2020) The global phosphorylation landscape of SARS-CoV-2 infection. Cell 182(3):685-712.e19",
"volume": "182",
"year": "2020"
},
{
"DOI": "10.1042/BCJ20160170",
"author": "JM Backer",
"doi-asserted-by": "publisher",
"first-page": "2251",
"issue": "15",
"journal-title": "Biochem J",
"key": "1869_CR93",
"unstructured": "Backer JM (2016) The intricate regulation and complex functions of the Class III phosphoinositide 3-kinase Vps34. Biochem J 473(15):2251–2271",
"volume": "473",
"year": "2016"
},
{
"DOI": "10.1371/journal.ppat.1007530",
"author": "Z Feng",
"doi-asserted-by": "publisher",
"first-page": "e1007530",
"issue": "1",
"journal-title": "PLoS Path",
"key": "1869_CR94",
"unstructured": "Feng Z, Xu K, Kovalev N, Nagy PD (2019) Recruitment of Vps34 PI3K and enrichment of PI3P phosphoinositide in the viral replication compartment is crucial for replication of a positive-strand RNA virus. PLoS Path 15(1):e1007530",
"volume": "15",
"year": "2019"
},
{
"DOI": "10.1074/jbc.M114.582999",
"author": "JM Liefhebber",
"doi-asserted-by": "publisher",
"first-page": "21276",
"issue": "31",
"journal-title": "J Biol Chem",
"key": "1869_CR95",
"unstructured": "Liefhebber JM, Hague CV, Zhang Q, Wakelam MJ, McLauchlan J (2014) Modulation of triglyceride and cholesterol ester synthesis impairs assembly of infectious hepatitis C virus. J Biol Chem 289(31):21276–21288",
"volume": "289",
"year": "2014"
},
{
"DOI": "10.1101/2020.07.18.210211",
"doi-asserted-by": "crossref",
"key": "1869_CR96",
"unstructured": "Silvas Jesus A, Jureka Alexander S, Nicolini Anthony M, Chvatal Stacie A, Basler Christopher F (2020) Inhibitors of VPS34 and lipid metabolism suppress SARS-CoV-2 replication. bioRxiv:2020.07.18.210211"
},
{
"DOI": "10.1002/jmv.26583",
"author": "CK Yuen",
"doi-asserted-by": "publisher",
"first-page": "2076",
"issue": "4",
"journal-title": "J Med Virol",
"key": "1869_CR97",
"unstructured": "Yuen CK, Wong WM, Mak LF, Wang X, Chu H, Yuen KY et al (2021) Suppression of SARS-CoV-2 infection in ex-vivo human lung tissues by targeting class III phosphoinositide 3-kinase. J Med Virol 93(4):2076–2083",
"volume": "93",
"year": "2021"
},
{
"DOI": "10.1371/journal.pbio.3000715",
"author": "EJ Snijder",
"doi-asserted-by": "publisher",
"first-page": "e3000715",
"issue": "6",
"journal-title": "PLoS Biol",
"key": "1869_CR98",
"unstructured": "Snijder EJ, Limpens RWAL, de Wilde AH, de Jong AWM, Zevenhoven-Dobbe JC, Maier HJ et al (2020) A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol 18(6):e3000715",
"volume": "18",
"year": "2020"
},
{
"DOI": "10.1016/j.chom.2010.05.013",
"author": "F Reggiori",
"doi-asserted-by": "publisher",
"first-page": "500",
"issue": "6",
"journal-title": "Cell Host Microbe",
"key": "1869_CR99",
"unstructured": "Reggiori F, Monastyrska I, Verheije MH, Calì T, Ulasli M, Bianchi S et al (2010) Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 7(6):500–508",
"volume": "7",
"year": "2010"
},
{
"DOI": "10.1038/s41467-019-13659-4",
"author": "NC Gassen",
"doi-asserted-by": "publisher",
"first-page": "5770",
"issue": "1",
"journal-title": "Nat Commun",
"key": "1869_CR100",
"unstructured": "Gassen NC, Niemeyer D, Muth D, Corman VM, Martinelli S, Gassen A et al (2019) SKP2 attenuates autophagy through beclin1-ubiquitination and its inhibition reduces MERS-coronavirus infection. Nat Commun 10(1):5770",
"volume": "10",
"year": "2019"
},
{
"DOI": "10.1038/ncb3053",
"author": "WE Dowdle",
"doi-asserted-by": "publisher",
"first-page": "1069",
"issue": "11",
"journal-title": "Nat Cell Biol",
"key": "1869_CR101",
"unstructured": "Dowdle WE, Nyfeler B, Nagel J, Elling RA, Liu S, Triantafellow E et al (2014) Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol 16(11):1069–1079",
"volume": "16",
"year": "2014"
},
{
"DOI": "10.15252/embj.2018100156",
"author": "L Su",
"doi-asserted-by": "publisher",
"first-page": "e100156",
"issue": "8",
"journal-title": "EMBO J",
"key": "1869_CR102",
"unstructured": "Su L, Zhou L, Chen FJ, Wang H, Qian H, Sheng Y et al (2019) Cideb controls sterol-regulated ER export of SREBP/SCAP by promoting cargo loading at ER exit sites. EMBO J 38(8):e100156",
"volume": "38",
"year": "2019"
},
{
"DOI": "10.1038/s41580-019-0190-7",
"author": "J Luo",
"doi-asserted-by": "publisher",
"first-page": "225",
"issue": "4",
"journal-title": "Nat Rev Mol Cell Biol",
"key": "1869_CR103",
"unstructured": "Luo J, Yang H, Song BL (2020) Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol 21(4):225–245",
"volume": "21",
"year": "2020"
},
{
"DOI": "10.1016/S0021-9258(18)50079-X",
"author": "DE Jones",
"doi-asserted-by": "publisher",
"first-page": "23216",
"issue": "32",
"journal-title": "J Biol Chem",
"key": "1869_CR104",
"unstructured": "Jones DE, Bevins CL (1992) Paneth cells of the human small intestine express an antimicrobial peptide gene. J Biol Chem 267(32):23216–23225",
"volume": "267",
"year": "1992"
},
{
"DOI": "10.1128/AAC.49.1.269-275.2005",
"author": "B Ericksen",
"doi-asserted-by": "publisher",
"first-page": "269",
"issue": "1",
"journal-title": "Antimicrob Agents Chemother",
"key": "1869_CR105",
"unstructured": "Ericksen B, Wu Z, Lu W, Lehrer RI (2005) Antibacterial activity and specificity of the six human {alpha}-defensins. Antimicrob Agents Chemother 49(1):269–275",
"volume": "49",
"year": "2005"
},
{
"DOI": "10.1021/jm501824a",
"author": "C Wang",
"doi-asserted-by": "publisher",
"first-page": "3083",
"issue": "7",
"journal-title": "J Med Chem",
"key": "1869_CR106",
"unstructured": "Wang C, Shen M, Gohain N, Tolbert WD, Chen F, Zhang N et al (2015) Design of a potent antibiotic peptide based on the active region of human defensin 5. J Med Chem 58(7):3083–3093",
"volume": "58",
"year": "2015"
},
{
"DOI": "10.1053/j.gastro.2020.05.015",
"author": "C Wang",
"doi-asserted-by": "publisher",
"first-page": "1145",
"issue": "3",
"journal-title": "Gastroenterology",
"key": "1869_CR107",
"unstructured": "Wang C, Wang S, Li D, Wei DQ, Zhao J, Wang J (2020) Human intestinal defensin 5 inhibits SARS-CoV-2 invasion by cloaking ACE2. Gastroenterology 159(3):1145–7.e4",
"volume": "159",
"year": "2020"
},
{
"DOI": "10.1016/j.mehy.2020.110244",
"author": "Y Niv",
"doi-asserted-by": "publisher",
"first-page": "110244",
"journal-title": "Med Hypotheses",
"key": "1869_CR108",
"unstructured": "Niv Y (2020) Defensin 5 for prevention of SARS-CoV-2 invasion and Covid-19 disease. Med Hypotheses 143:110244",
"volume": "143",
"year": "2020"
},
{
"DOI": "10.1016/0092-8674(93)90327-M",
"author": "D Sako",
"doi-asserted-by": "publisher",
"first-page": "1179",
"issue": "6",
"journal-title": "Cell",
"key": "1869_CR109",
"unstructured": "Sako D, Chang XJ, Barone KM, Vachino G, White HM, Shaw G et al (1993) Expression cloning of a functional glycoprotein ligand for P-selectin. Cell 75(6):1179–1186",
"volume": "75",
"year": "1993"
},
{
"DOI": "10.1016/S0092-8674(00)00138-0",
"author": "WS Somers",
"doi-asserted-by": "publisher",
"first-page": "467",
"issue": "3",
"journal-title": "Cell",
"key": "1869_CR110",
"unstructured": "Somers WS, Tang J, Shaw GD, Camphausen RT (2000) Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1. Cell 103(3):467–479",
"volume": "103",
"year": "2000"
},
{
"DOI": "10.1038/nm.1961",
"author": "Y Nishimura",
"doi-asserted-by": "publisher",
"first-page": "794",
"issue": "7",
"journal-title": "Nat Med",
"key": "1869_CR111",
"unstructured": "Nishimura Y, Shimojima M, Tano Y, Miyamura T, Wakita T, Shimizu H (2009) Human P-selectin glycoprotein ligand-1 is a functional receptor for enterovirus 71. Nat Med 15(7):794–797",
"volume": "15",
"year": "2009"
},
{
"DOI": "10.1038/s41564-019-0372-2",
"author": "Y Liu",
"doi-asserted-by": "publisher",
"first-page": "813",
"issue": "5",
"journal-title": "Nat Microbiol",
"key": "1869_CR112",
"unstructured": "Liu Y, Fu Y, Wang Q, Li M, Zhou Z, Dabbagh D et al (2019) Proteomic profiling of HIV-1 infection of human CD4 T cells identifies PSGL-1 as an HIV restriction factor. Nat Microbiol 4(5):813–825",
"volume": "4",
"year": "2019"
},
{
"key": "1869_CR113",
"unstructured": "He S, Hetrick B, Dabbagh D, Akhrymuk IV, Kehn-Hall K, Freed EO, et al (2020) PSGL-1 blocks SARS-CoV-2 S protein-mediated virus attachment and infection of target cells. bioRxiv:2020.05.01.073387"
},
{
"DOI": "10.1073/pnas.1916054117",
"author": "Y Fu",
"doi-asserted-by": "publisher",
"first-page": "9537",
"issue": "17",
"journal-title": "Proc Natl Acad Sci USA",
"key": "1869_CR114",
"unstructured": "Fu Y, He S, Waheed AA, Dabbagh D, Zhou Z, Trinité B et al (2020) PSGL-1 restricts HIV-1 infectivity by blocking virus particle attachment to target cells. Proc Natl Acad Sci USA 117(17):9537–9545",
"volume": "117",
"year": "2020"
},
{
"DOI": "10.1073/pnas.1712592114",
"author": "W Li",
"doi-asserted-by": "publisher",
"first-page": "E8508",
"issue": "40",
"journal-title": "Proc Natl Acad Sci USA",
"key": "1869_CR115",
"unstructured": "Li W, Hulswit RJG, Widjaja I, Raj VS, McBride R, Peng W et al (2017) Identification of sialic acid-binding function for the middle east respiratory syndrome coronavirus spike glycoprotein. Proc Natl Acad Sci USA 114(40):E8508–E8517",
"volume": "114",
"year": "2017"
},
{
"DOI": "10.1016/j.chom.2020.08.004",
"author": "P Zhao",
"doi-asserted-by": "publisher",
"first-page": "586",
"issue": "4",
"journal-title": "Cell Host Microbe",
"key": "1869_CR116",
"unstructured": "Zhao P, Praissman JL, Grant OC, Cai Y, Xiao T, Rosenbalm KE et al (2020) Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe 28(4):586-601.e6",
"volume": "28",
"year": "2020"
},
{
"DOI": "10.1016/j.chom.2020.05.008",
"author": "A Park",
"doi-asserted-by": "publisher",
"first-page": "870",
"issue": "6",
"journal-title": "Cell Host Microbe",
"key": "1869_CR117",
"unstructured": "Park A, Iwasaki A (2020) Type I and type III interferons—induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe 27(6):870–878",
"volume": "27",
"year": "2020"
},
{
"DOI": "10.1016/j.immuni.2012.11.005",
"author": "SY Liu",
"doi-asserted-by": "publisher",
"first-page": "92",
"issue": "1",
"journal-title": "Immunity",
"key": "1869_CR118",
"unstructured": "Liu SY, Aliyari R, Chikere K, Li G, Marsden MD, Smith JK et al (2013) Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity 38(1):92–105",
"volume": "38",
"year": "2013"
},
{
"DOI": "10.1074/jbc.273.51.34316",
"author": "EG Lund",
"doi-asserted-by": "publisher",
"first-page": "34316",
"issue": "51",
"journal-title": "J Biol Chem",
"key": "1869_CR119",
"unstructured": "Lund EG, Kerr TA, Sakai J, Li WP, Russell DW (1998) cDNA cloning of mouse and human cholesterol 25-hydroxylases, polytopic membrane proteins that synthesize a potent oxysterol regulator of lipid metabolism. J Biol Chem 273(51):34316–34327",
"volume": "273",
"year": "1998"
},
{
"DOI": "10.1016/j.immuni.2017.02.012",
"author": "C Li",
"doi-asserted-by": "publisher",
"first-page": "446",
"issue": "3",
"journal-title": "Immunity",
"key": "1869_CR120",
"unstructured": "Li C, Deng YQ, Wang S, Ma F, Aliyari R, Huang XY et al (2017) 25-Hydroxycholesterol protects host against zika virus infection and its associated microcephaly in a mouse model. Immunity 46(3):446–456",
"volume": "46",
"year": "2017"
},
{
"DOI": "10.15252/embj.2020106057",
"author": "S Wang",
"doi-asserted-by": "publisher",
"first-page": "6057",
"issue": "21",
"journal-title": "EMBO J",
"key": "1869_CR121",
"unstructured": "Wang S, Li W, Hui H, Tiwari SK, Zhang Q, Croker BA et al (2020) Cholesterol 25-hydroxylase inhibits SARS-CoV-2 and other coronaviruses by depleting membrane cholesterol. EMBO J 39(21):6057",
"volume": "39",
"year": "2020"
},
{
"DOI": "10.1074/jbc.RA119.010393",
"author": "KA Johnson",
"doi-asserted-by": "publisher",
"first-page": "17289",
"issue": "46",
"journal-title": "J Biol Chem",
"key": "1869_CR122",
"unstructured": "Johnson KA, Endapally S, Vazquez DC, Infante RE, Radhakrishnan A (2019) Ostreolysin A and anthrolysin O use different mechanisms to control movement of cholesterol from the plasma membrane to the endoplasmic reticulum. J Biol Chem 294(46):17289–17300",
"volume": "294",
"year": "2019"
},
{
"DOI": "10.1038/s41564-020-0701-5",
"author": "ME Abrams",
"doi-asserted-by": "publisher",
"first-page": "929",
"issue": "7",
"journal-title": "Nat Microbiol",
"key": "1869_CR123",
"unstructured": "Abrams ME, Johnson KA, Perelman SS, Zhang LS, Endapally S, Mar KB et al (2020) Oxysterols provide innate immunity to bacterial infection by mobilizing cell surface accessible cholesterol. Nat Microbiol 5(7):929–942",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.3390/v11111020",
"author": "J Yu",
"doi-asserted-by": "publisher",
"first-page": "1020",
"issue": "11",
"journal-title": "Viruses",
"key": "1869_CR124",
"unstructured": "Yu J, Liu SL (2019) Emerging role of LY6E in virus-host interactions. Viruses 11(11):1020",
"volume": "11",
"year": "2019"
},
{
"DOI": "10.1128/JVI.00562-20",
"author": "X Zhao",
"doi-asserted-by": "publisher",
"first-page": "e00562",
"issue": "18",
"journal-title": "J Virol",
"key": "1869_CR125",
"unstructured": "Zhao X, Zheng S, Chen D, Zheng M, Li X, Li G et al (2020) LY6E restricts entry of human coronaviruses, including currently pandemic SARS-CoV-2. J Virol 94(18):e00562-e620",
"volume": "94",
"year": "2020"
},
{
"DOI": "10.1038/s41467-018-06000-y",
"author": "KB Mar",
"doi-asserted-by": "publisher",
"first-page": "3603",
"issue": "1",
"journal-title": "Nat Commun",
"key": "1869_CR126",
"unstructured": "Mar KB, Rinkenberger NR, Boys IN, Eitson JL, McDougal MB, Richardson RB et al (2018) LY6E mediates an evolutionarily conserved enhancement of virus infection by targeting a late entry step. Nat Commun 9(1):3603",
"volume": "9",
"year": "2018"
},
{
"DOI": "10.1038/s41564-020-0769-y",
"author": "S Pfaender",
"doi-asserted-by": "publisher",
"first-page": "1330",
"issue": "11",
"journal-title": "Nat Microbiol",
"key": "1869_CR127",
"unstructured": "Pfaender S, Mar KB, Michailidis E, Kratzel A, BoysV’kovski INP et al (2020) LY6E impairs coronavirus fusion and confers immune control of viral disease. Nat Microbiol 5(11):1330–1339",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.1073/pnas.1913232116",
"author": "JL Meagher",
"doi-asserted-by": "publisher",
"first-page": "24303",
"issue": "48",
"journal-title": "Proc Natl Acad Sci USA",
"key": "1869_CR128",
"unstructured": "Meagher JL, Takata M, Gonçalves-Carneiro D, Keane SC, Rebendenne A, Ong H et al (2019) Structure of the zinc-finger antiviral protein in complex with RNA reveals a mechanism for selective targeting of CG-rich viral sequences. Proc Natl Acad Sci USA 116(48):24303–24309",
"volume": "116",
"year": "2019"
},
{
"DOI": "10.1128/JVI.01337-19",
"author": "M Ficarelli",
"doi-asserted-by": "publisher",
"first-page": "e01337",
"issue": "6",
"journal-title": "J Virol",
"key": "1869_CR129",
"unstructured": "Ficarelli M, Antzin-Anduetza I, Hugh-White R, Firth AE, Sertkaya H, Wilson H et al (2020) CpG dinucleotides inhibit HIV-1 replication through zinc finger antiviral protein (ZAP)-dependent and -independent mechanisms. J Virol 94(6):e01337-e1419",
"volume": "94",
"year": "2020"
},
{
"DOI": "10.1038/nature24039",
"author": "MA Takata",
"doi-asserted-by": "publisher",
"first-page": "124",
"issue": "7674",
"journal-title": "Nature",
"key": "1869_CR130",
"unstructured": "Takata MA, Gonçalves-Carneiro D, Zang TM, Soll SJ, York A, Blanco-Melo D et al (2017) CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature 550(7674):124–127",
"volume": "550",
"year": "2017"
},
{
"DOI": "10.1016/j.virol.2007.08.010",
"author": "PC Woo",
"doi-asserted-by": "publisher",
"first-page": "431",
"issue": "2",
"journal-title": "Virology",
"key": "1869_CR131",
"unstructured": "Woo PC, Wong BH, Huang Y, Lau SK, Yuen KY (2007) Cytosine deamination and selection of CpG suppressed clones are the two major independent biological forces that shape codon usage bias in coronaviruses. Virology 369(2):431–442",
"volume": "369",
"year": "2007"
},
{
"DOI": "10.1093/molbev/msaa094",
"author": "X Xia",
"doi-asserted-by": "publisher",
"first-page": "2699",
"issue": "9",
"journal-title": "Mol Biol Evol",
"key": "1869_CR132",
"unstructured": "Xia X (2020) Extreme genomic CpG deficiency in SARS-CoV-2 and evasion of host antiviral defense. Mol Biol Evol 37(9):2699–2705",
"volume": "37",
"year": "2020"
},
{
"DOI": "10.1128/mBio.01930-20",
"author": "R Nchioua",
"doi-asserted-by": "publisher",
"first-page": "e01930",
"issue": "5",
"journal-title": "mBio",
"key": "1869_CR133",
"unstructured": "Nchioua R, Kmiec D, Müller JA, Conzelmann C, Groß R, Swanson CM et al (2020) SARS-CoV-2 is restricted by zinc finger antiviral protein despite preadaptation to the low-CpG environment in humans. mBio 11(5):e01930-20",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1002/iub.379",
"author": "NB Calcaterra",
"doi-asserted-by": "publisher",
"first-page": "707",
"issue": "10",
"journal-title": "IUBMB Life",
"key": "1869_CR134",
"unstructured": "Calcaterra NB, Armas P, Weiner AM, Borgognone M (2010) CNBP: a multifunctional nucleic acid chaperone involved in cell death and proliferation control. IUBMB Life 62(10):707–714",
"volume": "62",
"year": "2010"
},
{
"DOI": "10.1038/s41564-020-00846-z",
"author": "N Schmidt",
"doi-asserted-by": "publisher",
"first-page": "339",
"issue": "3",
"journal-title": "Nat Microbiol",
"key": "1869_CR135",
"unstructured": "Schmidt N, Lareau CA, Keshishian H, Ganskih S, Schneider C, Hennig T et al (2020) The SARS-CoV-2 RNA-protein interactome in infected human cells. Nat Microbiol 6(3):339–353",
"volume": "6",
"year": "2020"
},
{
"DOI": "10.1371/journal.pgen.0040021",
"author": "JA Kerns",
"doi-asserted-by": "publisher",
"first-page": "e21",
"issue": "1",
"journal-title": "PLoS Genet",
"key": "1869_CR136",
"unstructured": "Kerns JA, Emerman M, Malik HS (2008) Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet 4(1):e21",
"volume": "4",
"year": "2008"
},
{
"DOI": "10.1016/j.molcel.2020.08.006",
"author": "K Klann",
"doi-asserted-by": "publisher",
"first-page": "164",
"issue": "1",
"journal-title": "Mol Cell",
"key": "1869_CR137",
"unstructured": "Klann K, Bojkova D, Tascher G, Ciesek S, Münch C, Cinatl J (2020) Growth factor receptor signaling inhibition prevents SARS-CoV-2 replication. Mol Cell 80(1):164–74.e4",
"volume": "80",
"year": "2020"
},
{
"DOI": "10.1074/jbc.M114.621730",
"author": "BD Fonseca",
"doi-asserted-by": "publisher",
"first-page": "15996",
"issue": "26",
"journal-title": "J Biol Chem",
"key": "1869_CR138",
"unstructured": "Fonseca BD, Zakaria C, Jia JJ, Graber TE, Svitkin Y, Tahmasebi S et al (2015) La-related protein 1 (LARP1) represses terminal oligopyrimidine (TOP) mRNA translation downstream of mTOR complex 1 (mTORC1). J Biol Chem 290(26):15996–16020",
"volume": "290",
"year": "2015"
}
],
"reference-count": 138,
"references-count": 138,
"relation": {},
"resource": {
"primary": {
"URL": "https://link.springer.com/10.1007/s11262-021-01869-2"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Virology",
"Genetics",
"Molecular Biology",
"General Medicine"
],
"subtitle": [],
"title": "Host proviral and antiviral factors for SARS-CoV-2",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1007/springer_crossmark_policy",
"volume": "57"
}