Uncovering Overlapping Gene Networks and Potential Therapeutic Targets in Osteoporosis and COVID‐19 Through Bioinformatics Analysis
et al., International Journal of Endocrinology, doi:10.1155/ije/8816596, Jan 2025
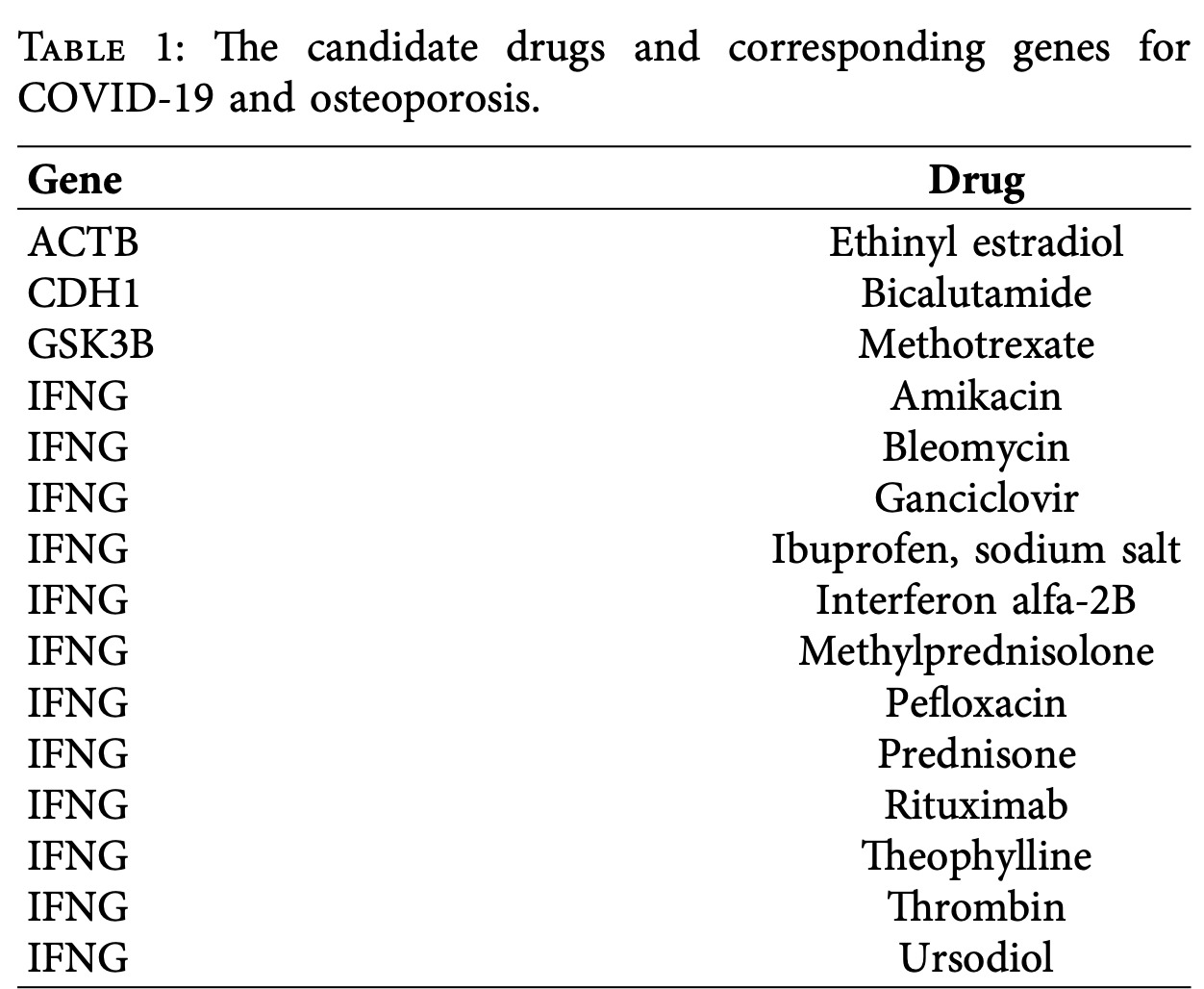
In silico bioinformatics study identifying 15 potential therapeutic agents for treating osteoporosis patients with COVID-19 through analysis of shared molecular pathways. Authors found that cytokine-cytokine receptor interactions and the Hippo signaling pathway are central to both conditions.
Luo et al., 31 Jan 2025, peer-reviewed, 6 authors.
Contact: xmleaf@126.com, wanzheng0626@xmu.edu.cn.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Uncovering Overlapping Gene Networks and Potential Therapeutic Targets in Osteoporosis and COVID‐19 Through Bioinformatics Analysis
International Journal of Endocrinology, doi:10.1155/ije/8816596
Background: Osteoporosis is a progressive bone disease characterized by reduced bone density and deterioration of bone microarchitecture, predominantly afecting the elderly population. Te ongoing COVID-19 pandemic has introduced additional challenges in osteoporosis management, potentially due to systemic infammation and direct viral impacts on bone metabolism. Tis study aims to identify common diferentially expressed genes (DEGs) and key molecular pathways shared between osteoporosis and COVID-19, with the goal of uncovering potential therapeutic targets through bioinformatics analysis. Methods: Publicly available gene expression datasets GSE164805 (osteoporosis) and GSE230665 (COVID-19) were analyzed to identify overlapping DEGs. Functional enrichment analysis using Gene Ontology (GO), pathway analysis, protein-protein interaction (PPI) network construction, and transcription factor (TF)-hub gene regulatory network analysis were performed to explore the biological signifcance and regulatory mechanisms of these DEGs. Results: A total of 325 common DEGs were identifed between osteoporosis and COVID-19. GO enrichment analysis revealed signifcant involvement in signal transduction and plasma membrane components. Pathway analysis highlighted the "cytokine-cytokine receptor interaction" pathway as a central player. PPI network analysis identifed a module of 193 genes with 397 interactions, from which 10 key hub genes were prioritized: ACTB, CDH1, RPS8, IFNG, RPL17, UBC, RPL36, RPS4Y1, GSK3B, and FGF13. Furthermore, 76 TFs were found to regulate these hub genes, and 15 existing drugs targeting four of these hub genes were identifed. Conclusion: Tis integrative bioinformatics study reveals 15 candidate therapeutic agents that target key regulatory genes shared between osteoporosis and COVID-19, ofering promising treatment strategies for osteoporotic patients, especially those impacted by or at risk of SARS-CoV-2 infection.
Ethics Statement Te datasets analyzed in this paper are publicly available and openly accessible. Since the data are not subject to individual privacy concerns and were collected without the involvement of the current researchers, ethical approval from a local ethics committee was not necessary.
Disclosure All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest Te authors declare no conficts of interest.
Author Contributions Zheng Wan and Yuwen Luo designed and coordinated the study. Yuwen Luo, Shizhen Liu, Xianyin Liu, Shu Zhong, and Ye Wang acquired and analyzed the data. All authors wrote the manuscript.
References
Amit, Beni, Biber, Grinberg, Leshem et al., Postvaccination COVID-19 Among Healthcare Workers, Israel, Emerging Infectious Diseases, doi:10.3201/eid2704.210016
Barrett, Wilhite, Ledoux, NCBI GEO: Archive for Functional Genomics Data Sets--Update, Nucleic Acids Research, doi:10.1093/nar/gks1193
Brown, Long-Term Treatment of Postmenopausal Osteoporosis, Endocrinol Metab (Seoul), doi:10.3803/enm.2021.301
Cannon, Stevenson, Stahl, DGIdb 5.0: Rebuilding the Drug-Gene Interaction Database for Precision Medicine and Drug Discovery Platforms, Nucleic Acids Research, doi:10.1093/nar/gkad1040
Carvelli, Demaria, Vély, Association of COVID-19 Infammation With Activation of the C5a-C5aR1 Axis, Nature, doi:10.1038/s41586-020-2600-6
Coughlan, Dockery, Osteoporosis and Fracture Risk in Older People, Clinical Medicine, doi:10.7861/clinmedicine.14-2-187
Fang, Chen, Liu, Age-Related GSK3β Overexpression Drives Podocyte Senescence and Glomerular Aging, Journal of Clinical Investigation, doi:10.1172/jci141848
Freshour, Kiwala, Cotto, Integration of the Drug-Gene Interaction Database (DGIdb 4.0) With Open Crowdsource Eforts, Nucleic Acids Research, doi:10.1093/nar/gkaa1084
Fu, Hu, Lan, Guan, Luo et al., Te Hippo Signalling Pathway and Its Implications in Human Health and Diseases, Signal Transduction and Targeted Terapy, doi:10.1038/s41392-022-01191-9
Gamble, Heller, Davis, Hereditary Difuse Gastric Cancer Syndrome and the Role of CDH1: A Review, JAMA Surgery, doi:10.1001/jamasurg.2020.6155
Garcia, Jeyachandran, Wang, Hippo Signaling Pathway Activation During SARS-CoV-2 Infection Contributes to Host Antiviral Response, PLoS Biology, doi:10.1371/journal.pbio.3001851
Gennari, Merlotti, Paola, Estrogen Receptor Gene Polymorphisms and the Genetics of Osteoporosis: A HuGE Review, American Journal of Epidemiology, doi:10.1093/aje/kwi055
Gong, Zhang, Dong, Integrated Bioinformatics Analysis for Identifcating the Terapeutic Targets of Aspirin in Small Cell Lung Cancer, Journal of Biomedical Informatics, doi:10.1016/j.jbi.2018.11.001
Guo, Liu, Wang, Sun, Greenaway, ACTB in Cancer, Clinica Chimica Acta, doi:10.1016/j.cca.2012.12.012
Hall, Foulkes, Insalata, Protection Against SARS-CoV-2 After Covid-19 Vaccination and Previous Infection, New England Journal of Medicine, doi:10.1056/nejmoa2118691
Hansford, Kaurah, Li-Chang, Hereditary Difuse Gastric Cancer Syndrome: CDH1 Mutations and Beyond, JAMA Oncology, doi:10.1001/jamaoncol.2014.168
Huang, Chen, Liu, CDK1/2/5 Inhibition Overcomes IFNG-Mediated Adaptive Immune Resistance in Pancreatic Cancer, Gut, doi:10.1136/gutjnl-2019-320441
Jafarzadeh, Nemati, Jafarzadeh, Contribution of STAT3 to the Pathogenesis of COVID-19, Microbial Pathogenesis, doi:10.1016/j.micpath.2021.104836
Lam, Duttke, Odish, Profling Transcription Initiation in Peripheral Leukocytes Reveals Severity-Associated Cis-Regulatory Elements in Critical COVID-19, bioRxiv, doi:10.1101/2021.08.24.457187
Li, Dong, Leier, Positive-Unlabeled Learning in Bioinformatics and Computational Biology: A Brief Review, Briefngs in Bioinformatics, doi:10.1093/bib/bbab461
Li, Samuel, Ul Haq, Characterizing the Oncogenic Importance and Exploring Gene-Immune Cells Correlation of ACTB in Human Cancers, American Journal of Cancer Research
Libuit, Doughty, Otieno, Accelerating Bioinformatics Implementation in Public Health, Microbial Genomics, doi:10.1099/mgen.0.001051
Liu, Verma, Garcia, Targeting the Coronavirus Nucleocapsid Protein Trough GSK-3 Inhibition, Proceedings of the National Academy of Sciences of the United States of America, doi:10.1073/pnas.2113401118
Lv, Kang, Wu, Fluorosis Increases the Risk of Postmenopausal Osteoporosis by Stimulating Interferon Γ, Biochemical and Biophysical Research Communications, doi:10.1016/j.bbrc.2016.09.083
Ma, Meng, Chen, Guan, Te Hippo Pathway: Biology and Pathophysiology, Annual Review of Biochemistry, doi:10.1146/annurev-biochem-013118-111829
Majumder, Minko, Recent Developments on Terapeutic and Diagnostic Approaches for COVID-19, Te AAPS Journal, doi:10.1208/s12248-020-00532-2
Rachner, Khosla, Hofbauer, Osteoporosis: now and the Future, Te Lancet, doi:10.1016/s0140-6736(10)62349-5
Rudd, GSK-3 Inhibition as a Terapeutic Approach Against Sars CoV2: Dual Beneft of Inhibiting Viral Replication While Potentiating the Immune Response, Frontiers in Immunology, doi:10.3389/fimmu.2020.01638
Sherman, Hao, Qiu, DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update), Nucleic Acids Research, doi:10.1093/nar/gkac194
Szklarczyk, Gable, Nastou, Te STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/ Measurement Sets, Nucleic Acids Research, doi:10.1093/nar/gkaa1074
Tu, Bao, MBL2 Genetic Polymorphisms and Severe COVID-19: A Protocol for Systematic Review and Meta-Analysis, Medicine (Baltimore), doi:10.1097/md.0000000000029405
Umakanthan, Sahu, Ranade, Origin, Transmission, Diagnosis and Management of Coronavirus Disease 2019 (COVID-19), Postgraduate Medical Journal, doi:10.1136/postgradmedj-2020-138234
Van Gelder, Hooft, Van Rijswijk, Bioinformatics in the Netherlands: Te Value of a Nationwide Community, Briefngs in Bioinformatics, doi:10.1093/bib/bbx087
Wan, Zou, Gao, Cdh1 Regulates Osteoblast Function Trough an APC/C-Independent Modulation of Smurf1, Molecular Cell, doi:10.1016/j.molcel.2011.09.024
Wooller, Benstead-Hume, Chen, Ali, Pearl, Bioinformatics in Translational Drug Discovery, Bioscience Reports, doi:10.1042/bsr20160180
Xiao, Wan, Liu, Screening of Terapeutic Targets for Pancreatic Cancer by Bioinformatics Methods, Hormone and Metabolic Research, doi:10.1055/a-2007-2715
Xu, Han, Li, Efective Treatment of Severe COVID-19 Patients With Tocilizumab, Proceedings of the National Academy of Sciences of the United States of America, doi:10.1073/pnas.2005615117
Yan, Cao, Sun, Sun, Gao, Inhibition of GSK3B Phosphorylation Improves Glucose and Lipid Metabolism Disorder, Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease, doi:10.1016/j.bbadis.2023.166726
Yan, Tang, Chen, Imperatorin Promotes Osteogenesis and Suppresses Osteoclast by Activating AKT/ GSK3 β/β-Catenin Pathways, Journal of Cellular and Molecular Medicine, doi:10.1111/jcmm.14915
Yang, Kuang, Kang, Zhao, Ma et al., Human Umbilical Cord Mesenchymal Stem Cell-Derived Exosomes Act via the miR-1263/Mob1/Hippo Signaling Pathway to Prevent Apoptosis in Disuse Osteoporosis, Biochemical and Biophysical Research Communications, doi:10.1016/j.bbrc.2020.02.001
Yu, Xia, Te Epidemiology of Osteoporosis, Associated Fragility Fractures, and Management Gap in China, Arch Osteoporos, doi:10.1007/s11657-018-0549-y
Zheng, Vrindts, Lopez, Increase in Cytokine Production (IL-1 Beta, IL-6, TNF-Alpha but Not IFN-Gamma, GM-CSF or LIF) by Stimulated Whole Blood Cells in Postmenopausal Osteoporosis, Maturitas, doi:10.1016/s0378-5122(96)01080-8
Zhou, Ren, Zhang, Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients, Cell Host & Microbe, doi:10.1016/j.chom.2020.04.017
Zhou, Soufan, Ewald, Hancock, Basu et al., NetworkAnalyst 3.0: A Visual Analytics Platform for Comprehensive Gene Expression Profling and Meta-Analysis, Nucleic Acids Research, doi:10.1093/nar/gkz240
Zhu, Zhang, Xiong, Fan, Li et al., Circ-GSK3B Up-Regulates GSK3B to Suppress the Progression of Lung Adenocarcinoma, Cancer Gene Terapy, doi:10.1038/s41417-022-00489-8
DOI record:
{
"DOI": "10.1155/ije/8816596",
"ISSN": [
"1687-8337",
"1687-8345"
],
"URL": "http://dx.doi.org/10.1155/ije/8816596",
"abstract": "<jats:p><jats:bold>Background:</jats:bold> Osteoporosis is a progressive bone disease characterized by reduced bone density and deterioration of bone microarchitecture, predominantly affecting the elderly population. The ongoing COVID‐19 pandemic has introduced additional challenges in osteoporosis management, potentially due to systemic inflammation and direct viral impacts on bone metabolism. This study aims to identify common differentially expressed genes (DEGs) and key molecular pathways shared between osteoporosis and COVID‐19, with the goal of uncovering potential therapeutic targets through bioinformatics analysis.</jats:p><jats:p><jats:bold>Methods:</jats:bold> Publicly available gene expression datasets GSE164805 (osteoporosis) and GSE230665 (COVID‐19) were analyzed to identify overlapping DEGs. Functional enrichment analysis using Gene Ontology (GO), pathway analysis, protein–protein interaction (PPI) network construction, and transcription factor (TF)–hub gene regulatory network analysis were performed to explore the biological significance and regulatory mechanisms of these DEGs.</jats:p><jats:p><jats:bold>Results:</jats:bold> A total of 325 common DEGs were identified between osteoporosis and COVID‐19. GO enrichment analysis revealed significant involvement in signal transduction and plasma membrane components. Pathway analysis highlighted the “cytokine–cytokine receptor interaction” pathway as a central player. PPI network analysis identified a module of 193 genes with 397 interactions, from which 10 key hub genes were prioritized: ACTB, CDH1, RPS8, IFNG, RPL17, UBC, RPL36, RPS4Y1, GSK3B, and FGF13. Furthermore, 76 TFs were found to regulate these hub genes, and 15 existing drugs targeting four of these hub genes were identified.</jats:p><jats:p><jats:bold>Conclusion:</jats:bold> This integrative bioinformatics study reveals 15 candidate therapeutic agents that target key regulatory genes shared between osteoporosis and COVID‐19, offering promising treatment strategies for osteoporotic patients, especially those impacted by or at risk of SARS‐CoV‐2 infection.</jats:p>",
"alternative-id": [
"10.1155/ije/8816596"
],
"article-number": "8816596",
"assertion": [
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2024-09-14"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "2025-08-19"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 3,
"value": "2025-08-30"
}
],
"author": [
{
"ORCID": "https://orcid.org/0009-0004-5898-9613",
"affiliation": [],
"authenticated-orcid": false,
"family": "Luo",
"given": "Yuwen",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0009-0000-1379-6173",
"affiliation": [],
"authenticated-orcid": false,
"family": "Liu",
"given": "Shizhen",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0009-0002-4404-4825",
"affiliation": [],
"authenticated-orcid": false,
"family": "Liu",
"given": "Xianyin",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0009-0003-5566-5608",
"affiliation": [],
"authenticated-orcid": false,
"family": "Zhong",
"given": "Shu",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0009-0006-7109-6144",
"affiliation": [],
"authenticated-orcid": false,
"family": "Wang",
"given": "Ye",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-8455-3478",
"affiliation": [],
"authenticated-orcid": false,
"family": "Wan",
"given": "Zheng",
"sequence": "additional"
}
],
"container-title": "International Journal of Endocrinology",
"container-title-short": "International Journal of Endocrinology",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"onlinelibrary.wiley.com"
]
},
"created": {
"date-parts": [
[
2025,
8,
30
]
],
"date-time": "2025-08-30T10:04:46Z",
"timestamp": 1756548286000
},
"deposited": {
"date-parts": [
[
2025,
8,
30
]
],
"date-time": "2025-08-30T10:04:48Z",
"timestamp": 1756548288000
},
"editor": [
{
"affiliation": [],
"family": "Saleem",
"given": "Suraiya",
"sequence": "additional"
}
],
"funder": [
{
"DOI": "10.13039/501100003392",
"award": [
"2022J05298"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100003392",
"id-type": "DOI"
}
],
"name": "Natural Science Foundation of Fujian Province"
}
],
"indexed": {
"date-parts": [
[
2025,
8,
30
]
],
"date-time": "2025-08-30T10:40:08Z",
"timestamp": 1756550408019,
"version": "3.44.0"
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2025,
1
]
]
},
"journal-issue": {
"issue": "1",
"published-print": {
"date-parts": [
[
2025,
1
]
]
}
},
"language": "en",
"license": [
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 241,
"start": {
"date-parts": [
[
2025,
8,
30
]
],
"date-time": "2025-08-30T00:00:00Z",
"timestamp": 1756512000000
}
}
],
"link": [
{
"URL": "https://onlinelibrary.wiley.com/doi/pdf/10.1155/ije/8816596",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "311",
"original-title": [],
"prefix": "10.1155",
"published": {
"date-parts": [
[
2025,
1
]
]
},
"published-online": {
"date-parts": [
[
2025,
8,
30
]
]
},
"published-print": {
"date-parts": [
[
2025,
1
]
]
},
"publisher": "Wiley",
"reference": [
{
"DOI": "10.3803/enm.2021.301",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_1_2"
},
{
"DOI": "10.7861/clinmedicine.14-2-187",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_2_2"
},
{
"DOI": "10.1007/s11657-018-0549-y",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_3_2"
},
{
"DOI": "10.1056/nejmoa2118691",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_4_2"
},
{
"DOI": "10.1208/s12248-020-00532-2",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_5_2"
},
{
"DOI": "10.1136/postgradmedj-2020-138234",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_6_2"
},
{
"DOI": "10.3201/eid2704.210016",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_7_2"
},
{
"DOI": "10.1093/bib/bbx087",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_8_2"
},
{
"DOI": "10.1042/bsr20160180",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_9_2"
},
{
"DOI": "10.1093/bib/bbab461",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_10_2"
},
{
"DOI": "10.1099/mgen.0.001051",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_11_2"
},
{
"DOI": "10.1016/j.jbi.2018.11.001",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_12_2"
},
{
"DOI": "10.1093/nar/gks1193",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_13_2"
},
{
"DOI": "10.1093/nar/gkac194",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_14_2"
},
{
"DOI": "10.1093/nar/gkaa1074",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_15_2"
},
{
"DOI": "10.1093/nar/gkz240",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_16_2"
},
{
"DOI": "10.1093/nar/gkaa1084",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_17_2"
},
{
"DOI": "10.1146/annurev-biochem-013118-111829",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_18_2"
},
{
"DOI": "10.1038/s41392-022-01191-9",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_19_2"
},
{
"DOI": "10.1016/j.bbrc.2020.02.001",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_20_2"
},
{
"DOI": "10.1371/journal.pbio.3001851",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_21_2"
},
{
"DOI": "10.1016/j.cca.2012.12.012",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_22_2"
},
{
"article-title": "Characterizing the Oncogenic Importance and Exploring Gene-Immune Cells Correlation of ACTB in Human Cancers",
"author": "Li G.",
"first-page": "758",
"journal-title": "American Journal of Cancer Research",
"key": "e_1_2_12_23_2",
"volume": "13",
"year": "2023"
},
{
"DOI": "10.1016/s0140-6736(10)62349-5",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_24_2"
},
{
"DOI": "10.1073/pnas.2005615117",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_25_2"
},
{
"DOI": "10.1001/jamaoncol.2014.168",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_26_2"
},
{
"DOI": "10.1001/jamasurg.2020.6155",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_27_2"
},
{
"DOI": "10.1016/j.molcel.2011.09.024",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_28_2"
},
{
"DOI": "10.1038/s41586-020-2600-6",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_29_2"
},
{
"DOI": "10.1172/jci141848",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_30_2"
},
{
"DOI": "10.1016/j.bbadis.2023.166726",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_31_2"
},
{
"DOI": "10.1038/s41417-022-00489-8",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_32_2"
},
{
"DOI": "10.1111/jcmm.14915",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_33_2"
},
{
"DOI": "10.1073/pnas.2113401118",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_34_2"
},
{
"DOI": "10.3389/fimmu.2020.01638",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_35_2"
},
{
"DOI": "10.1136/gutjnl-2019-320441",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_36_2"
},
{
"DOI": "10.1016/j.bbrc.2016.09.083",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_37_2"
},
{
"DOI": "10.1016/s0378-5122(96)01080-8",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_38_2"
},
{
"DOI": "10.1016/j.chom.2020.04.017",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_39_2"
},
{
"DOI": "10.1097/md.0000000000029405",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_40_2"
},
{
"DOI": "10.1093/aje/kwi055",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_41_2"
},
{
"DOI": "10.1016/j.micpath.2021.104836",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_42_2"
},
{
"DOI": "10.1101/2021.08.24.457187",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_43_2"
},
{
"DOI": "10.1093/nar/gkad1040",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_44_2"
},
{
"DOI": "10.1055/a-2007-2715",
"doi-asserted-by": "publisher",
"key": "e_1_2_12_45_2"
}
],
"reference-count": 45,
"references-count": 45,
"relation": {},
"resource": {
"primary": {
"URL": "https://onlinelibrary.wiley.com/doi/10.1155/ije/8816596"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Uncovering Overlapping Gene Networks and Potential Therapeutic Targets in Osteoporosis and COVID‐19 Through Bioinformatics Analysis",
"type": "journal-article",
"update-policy": "https://doi.org/10.1002/crossmark_policy",
"volume": "2025"
}