Monitoring SARS-CoV-2 Nsp13 helicase binding activity using expanded genetic code techniques
et al., RSC Chemical Biology, doi:10.1039/d4cb00230j, Apr 2025
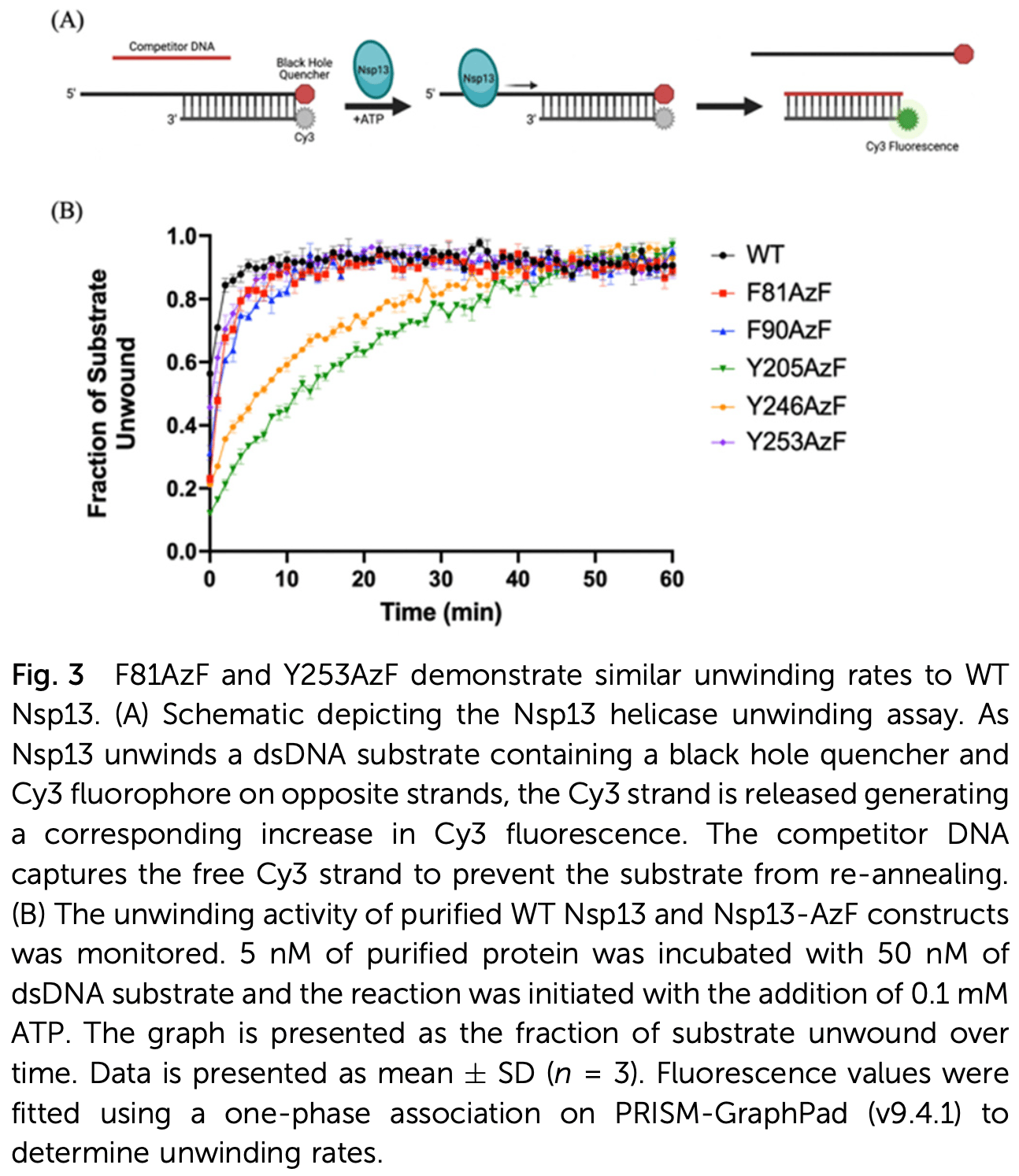
In vitro study showing that site-specific fluorescent labeling of SARS-CoV-2 Nsp13 helicase enables monitoring of its binding activity with nucleic acid substrates. Authors developed a genetic code expansion technique to incorporate p-azido-L-phenylalanine (AzF) at specific sites in Nsp13, allowing for fluorescent labeling with Cy5-dibenzocyclooctyne (DBCO). They identified two Nsp13-AzF constructs (F81AzF and Y253AzF) that maintained similar unwinding activity to wild-type Nsp13 while enabling efficient fluorescent labeling. Using Förster resonance energy transfer (FRET), they demonstrated that the F81AzF construct could monitor Nsp13 binding to nucleic acid substrates in a distance-dependent manner. This approach provides a novel method for screening potential Nsp13 inhibitors and studying the molecular mechanism of this essential viral helicase during SARS-CoV-2 replication.
Lundrigan et al., 21 Apr 2025, peer-reviewed, 4 authors.
Contact: john.pezacki@uottawa.ca.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Monitoring SARS-CoV-2 Nsp13 helicase binding activity using expanded genetic code techniques
RSC Chemical Biology, doi:10.1039/d4cb00230j
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) non-structural protein 13 (Nsp13) helicase is a multi-functional protein that can unwind dsDNA and dsRNA in an NTP-dependent manner. Given that this viral helicase is essential for viral replication and highly conserved among coronaviruses, a thorough understanding of the helicase's unwinding and binding activity may allow for the development of more effective pan-coronavirus therapeutics. Herein, we describe the use of genetic code expansion techniques to site-specifically incorporate the non-canonical amino acid (ncAA) p-azido-L-phenylalanine (AzF) into Nsp13 for fluorescent labelling of the enzyme with a conjugated Cy5 fluorophore. This Cy5labelled Nsp13-AzF can then be used in Fo ¨rster resonance energy transfer (FRET) experiments to investigate the dynamics of enzyme translocation on its substrate during binding and unwinding. Five sites (F81, F90, Y205, Y246, and Y253) were identified for AzF incorporation in Nsp13 and assessed for fluorescent labelling efficiency. The incorporation of AzF was confirmed to not interfere with the unwinding activity of the helicase. Subsequently, FRET-based binding assays were conducted to monitor the binding of Cy5-labelled Nsp13-AzF constructs to a series of fluorescently-labelled nucleic acid substrates in a distance-dependent manner. Overall, this approach not only allows for the direct monitoring of Nsp13's binding activity on its substrate, it may also introduce a novel method to screen for compounds that can inhibit this essential enzymatic activity during viral replication.
Author contributions C. Hum performed experimental work, data collection, analysis and writing the original draft. N. Ahmed contributed to conceptualization of the study. E. Lundrigan performed experimental work, data collection, and analysis and contributed to the writing and revising at the stage of review. J. P. Pezacki
Conflicts of interest There are no conflicts to declare.
References
Ablenas, Gidi, Powdrill, Hepatitis C Virus Helicase Binding Activity Monitored through Site-Specific Labeling Using an Expanded Genetic Code, ACS Infect. Dis, doi:10.1021/acsinfecdis.9b00220
Adedeji, Marchand, Velthuis, Mechanism of Nucleic Acid Unwinding by SARS-CoV Helicase, PLoS One, doi:10.1371/journal.pone.0036521
Adedeji, Singh, Calcaterra, Severe Acute Respiratory Syndrome Coronavirus Replication Inhibitor That Interferes with the Nucleic Acid Unwinding of the Viral Helicase, Antimicrob. Agents Chemother, doi:10.1128/AAC.00957-12
Berta, Badaoui, Martino, Modelling the active SARS-CoV-2 helicase complex as a basis for structurebased inhibitor design, Chem. Sci, doi:10.1039/D1SC02775A
Chen, Malone, Llewellyn, Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex, Cell, doi:10.1016/J.CELL.2020.07.033
Chin, Expanding and Reprogramming the Genetic Code of Cells and Animals, Annu. Rev. Biochem, doi:10.1146/annurev-biochem-060713-035737
Chin, Santoro, Martin, King, Wang et al., Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli, J. Am. Chem. Soc, doi:10.1021/ja027007w
Chin, Santoro, Martin, King, Wang et al., Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli, J. Am. Chem. Soc, doi:10.1021/ja027007w
Chudakov, Lukyanov, Lukyanov, Fluorescent proteins as a toolkit for in vivo imaging, Trends Biotechnol, doi:10.1016/j.tibtech.2005.10.005
Dong, Du, Gardner, An interactive web-based dashboard to track COVID-19 in real time, Published online, doi:10.1016/S1473-3099(20)30120-1
Gurung, In silico structure modelling of SARS-CoV-2 Nsp13 helicase and Nsp14 and repurposing of FDA approved antiviral drugs as dual inhibitors, Gene Rep, doi:10.1016/j.genrep.2020.100860
Ha, Single-molecule fluorescence methods for the study of nucleic acids, Curr. Opin. Struct. Biol, doi:10.1016/s0959-440x(00)00204-9
Herbert, Poptsova, Z-RNA and the Flipside of the SARS Nsp13 Helicase: Is There a Role for Flipons in Coronavirus-Induced Pathology?, Front. Immunol, doi:10.3389/fimmu.2022.912717
Hoogenboom, Thiol-Yne Chemistry: A Powerful Tool for Creating Highly Functional Materials, Angew. Chem., Int. Ed, doi:10.1002/anie.201000401
Hwang, Myong, Protein induced fluorescence enhancement (PIFE) for probing protein-nucleic acid interactions, Chem. Soc. Rev, doi:10.1039/c3cs60201j
Iqbal, Wang, Thompson, Lilley, Norman, The structure of cyanine 5 terminally attached to double-stranded DNA: Implications for FRET studies, Biochemistry, doi:10.1021/bi800773f
Ivanov, Thiel, Dobbe, Van Der Meer, Snijder et al., Multiple Enzymatic Activities Associated with Severe Acute Respiratory Syndrome Coronavirus Helicase, J. Virol, doi:10.1128/jvi.78.11.5619-5632.2004
Jakob, Gust, Grohmann, Evaluation and optimisation of unnatural amino acid incorporation and bioorthogonal bioconjugation for site-specific fluorescent labelling of proteins expressed in mammalian cells, Biochem. Biophys. Rep, doi:10.1016/j.bbrep.2018.10.011
Jang, Jeong, Kang, Sp, Yang et al., A high ATP concentration enhances the cooperative translocation of the SARS coronavirus helicase nsP13 in the unwinding of duplex RNA, Sci. Rep, doi:10.1038/s41598-020-61432-1
Jia, Yan, Ren, Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis, Nucleic Acids Res, doi:10.1093/nar/gkz409
Lang, Chin, Cellular incorporation of unnatural amino acids and bioorthogonal Labeling of Proteins, Chem. Rev, doi:10.1021/cr400355w
Lehmann, Snijder, Posthuma, Gorbalenya, What we know but do not understand about nidovirus helicases, Virus Res, doi:10.1016/j.virusres.2014.12.001
Martin, Li, Parvangada, Genetic conservation of SARS-CoV-2 RNA replication complex in globally circulating isolates and recently emerged variants from humans and minks suggests minimal pre-existing resistance to remdesivir, Antiviral Res, doi:10.1016/j.antiviral.2021.105033
Marx, Mickolajczyk, Craig, Thomas, Pfeffer et al., Observing inhibition of the SARS-CoV-2 helicase at single-nucleotide resolution, Nucleic Acids Res, doi:10.1093/nar/gkad660
Mathieu, Ritchie, Rode ´s-Guirao, Coronavirus Pandemic (COVID-19). Our World in Data
Mckay, Finn, Click Chemistry in Complex Mixtures: Bioorthogonal Bioconjugation, Chem. Biol, doi:10.1016/j.chembiol.2014.09.002
Mickolajczyk, Shelton, Grasso, Force-dependent stimulation of RNA unwinding by SARS-CoV-2 nsp13 helicase, Biophys. J, doi:10.1016/j.bpj.2020.11.2276
Newman, Douangamath, Yadzani, Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase, Nat. Commun, doi:10.1038/s41467-021-25166-6
Pak, Adegboye, Adekunle, Rahman, Mcbryde et al., Economic Consequences of the COVID-19 Outbreak: the Need for Epidemic Preparedness, Front. Public Health, doi:10.3389/fpubh.2020.00241
Park, Osinski, Hernandez, The mechanism of RNA capping by SARS-CoV-2, Nature, doi:10.1038/s41586-022-05185-z
Perez-Lemus, Mene ´ndez, Alvarado, Byle ´hn, De, Toward wide-spectrum antivirals against coronaviruses: Molecular characterization of SARS-CoV-2 NSP13 helicase inhibitors, Sci. Adv, doi:10.1126/sciadv.abj4526
Rosas-Lemus, Minasov, Shuvalova, Highresolution structures of the SARS-CoV-2 2 0 -O-methyltransferase reveal strategies for structure-based inhibitor design, Sci. Signaling, doi:10.1126/scisignal.abe1202
Roy, Hohng, Ha, A practical guide to singlemolecule FRET, Nat. Methods, doi:10.1038/nmeth.1208
Seybert, Hegyi, Siddell, Ziebuhr, The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5 0 -to-3 0 polarity, RNA, doi:10.1017/s1355838200000728
Shrestha, Jenei, Nagy, Vereb, Szo ¨ll+ osi, Understanding FRET as a research tool for cellular studies, Int. J. Mol. Sci, doi:10.3390/ijms16046718
Sun, Schultz, Kim, Therapeutic applications of an expanded genetic code, ChemBioChem, doi:10.1002/cbic.201402154
Tang, Comish, Kang, The hallmarks of COVID-19 disease, PLoS Pathog, doi:10.1371/journal.ppat.1008536
Tanner, Watt, Chai, The severe acute respiratory syndrome (SARS) coronavirus NTPasefhelicase belongs to a distinct class of 5 0 to 3 0 viral helicases, J. Biol. Chem, doi:10.1074/jbc.c300328200
Toseland, Fluorescent labeling and modification of proteins, J. Chem. Biol, doi:10.1007/s12154-013-0094-5
Viswanathan, Arya, Chan, Structural basis of RNA cap modification by SARS-CoV-2, Nat. Commun, doi:10.1038/s41467-020-17496-8
Wang, Sachdeva, Cox, Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET, Nat. Chem, doi:10.1038/nchem.1919
Wilamowski, Sherrell, Minasov, 2 0 -O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography, Proc. Natl. Acad. Sci. U. S. A, doi:10.1073/pnas.2100170118
Wu, Zhao, Yu, A new coronavirus associated with human respiratory disease in China, Nature, doi:10.1038/s41586-020-2008-3
Yan, Zhang, Ge, Architecture of a SARS-CoV-2 mini replication and transcription complex, Nat. Commun, doi:10.1038/s41467-020-19770-1
Yan, Zheng, Zeng, He, Cheng, Structural biology of SARS-CoV-2: open the door for novel therapies, Signal Transduction Targeted Ther, doi:10.1038/s41392-022-00884-5
Yu, Lee, Lee, Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13, Bioorg. Med. Chem. Lett, doi:10.1016/J.BMCL.2012.04.081
Zander, Holzmeister, Klose, Tinnefeld, Grohmann, Single-molecule FRET supports the two-state model of Argonaute action, RNA Biol, doi:10.4161/RNA.27446
Zeng, Weissmann, Bertolin, Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp13 helicase, Biochem. J, doi:10.1042/bcj20210201
Zhang, Li, Cowley, The nsp1, nsp13, and M Proteins Contribute to the Hepatotropism of Murine Coronavirus JHM.WU, J. Virol, doi:10.1128/jvi.03535-14
DOI record:
{
"DOI": "10.1039/d4cb00230j",
"ISSN": [
"2633-0679"
],
"URL": "http://dx.doi.org/10.1039/d4cb00230j",
"abstract": "<jats:p>The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) non-structural protein 13 (Nsp13) helicase is a multi-functional protein that can unwind dsDNA and dsRNA in an NTP-dependent manner.</jats:p>",
"author": [
{
"ORCID": "https://orcid.org/0000-0001-5896-9752",
"affiliation": [
{
"name": "Department of Chemistry and Biomolecular Sciences, University of Ottawa, Ottawa, Ontario K1N 6N5, Canada"
}
],
"authenticated-orcid": false,
"family": "Lundrigan",
"given": "Eryn",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Department of Chemistry and Biomolecular Sciences, University of Ottawa, Ottawa, Ontario K1N 6N5, Canada"
}
],
"family": "Hum",
"given": "Christine",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Chemistry and Biomolecular Sciences, University of Ottawa, Ottawa, Ontario K1N 6N5, Canada"
}
],
"family": "Ahmed",
"given": "Nadine",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-4233-8945",
"affiliation": [
{
"name": "Department of Chemistry and Biomolecular Sciences, University of Ottawa, Ottawa, Ontario K1N 6N5, Canada"
}
],
"authenticated-orcid": false,
"family": "Pezacki",
"given": "John Paul",
"sequence": "additional"
}
],
"container-title": "RSC Chemical Biology",
"container-title-short": "RSC Chem. Biol.",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"rsc.org"
]
},
"created": {
"date-parts": [
[
2025,
4,
21
]
],
"date-time": "2025-04-21T17:41:06Z",
"timestamp": 1745257266000
},
"deposited": {
"date-parts": [
[
2025,
4,
29
]
],
"date-time": "2025-04-29T08:31:19Z",
"timestamp": 1745915479000
},
"funder": [
{
"DOI": "10.13039/501100000038",
"award": [
"210719"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100000038",
"id-type": "DOI"
}
],
"name": "Natural Sciences and Engineering Research Council of Canada"
}
],
"indexed": {
"date-parts": [
[
2025,
4,
29
]
],
"date-time": "2025-04-29T09:10:09Z",
"timestamp": 1745917809898,
"version": "3.40.4"
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2025
]
]
},
"language": "en",
"license": [
{
"URL": "http://creativecommons.org/licenses/by-nc/3.0/",
"content-version": "vor",
"delay-in-days": 110,
"start": {
"date-parts": [
[
2025,
4,
21
]
],
"date-time": "2025-04-21T00:00:00Z",
"timestamp": 1745193600000
}
}
],
"link": [
{
"URL": "http://pubs.rsc.org/en/content/articlepdf/2025/CB/D4CB00230J",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "292",
"original-title": [],
"prefix": "10.1039",
"published": {
"date-parts": [
[
2025
]
]
},
"published-online": {
"date-parts": [
[
2025
]
]
},
"publisher": "Royal Society of Chemistry (RSC)",
"reference": [
{
"DOI": "10.1038/s41586-020-2008-3",
"author": "Wu",
"doi-asserted-by": "publisher",
"first-page": "265",
"issue": "7798",
"journal-title": "Nature",
"key": "D4CB00230J/cit1/1",
"volume": "579",
"year": "2020"
},
{
"DOI": "10.3389/fpubh.2020.00241",
"author": "Pak",
"doi-asserted-by": "publisher",
"first-page": "241",
"journal-title": "Front. Public Health",
"key": "D4CB00230J/cit2/1",
"volume": "8",
"year": "2020"
},
{
"DOI": "10.1371/journal.ppat.1008536",
"author": "Tang",
"doi-asserted-by": "publisher",
"first-page": "e1008536",
"issue": "5",
"journal-title": "PLoS Pathog.",
"key": "D4CB00230J/cit3/1",
"volume": "16",
"year": "2020"
},
{
"DOI": "10.1016/S1473-3099(20)30120-1",
"author": "Dong",
"doi-asserted-by": "publisher",
"key": "D4CB00230J/cit4/1",
"unstructured": "E.Dong , H.Du and L.Gardner An interactive web-based dashboard to track COVID-19 in real time . Published online 2020 10.1016/S1473-3099(20)30120-1",
"year": "2020"
},
{
"author": "Mathieu",
"key": "D4CB00230J/cit5/1",
"unstructured": "E.Mathieu , H.Ritchie , L.Rodés-Guirao , et al. Coronavirus Pandemic (COVID-19) . Our World in Data . Published online March 5, 2020. Accessed January 15, 2023. https://ourworldindata.org/coronavirus"
},
{
"DOI": "10.1128/jvi.03535-14",
"author": "Zhang",
"doi-asserted-by": "publisher",
"first-page": "3598",
"issue": "7",
"journal-title": "J. Virol.",
"key": "D4CB00230J/cit6/1",
"volume": "89",
"year": "2015"
},
{
"DOI": "10.1016/j.virusres.2014.12.001",
"author": "Lehmann",
"doi-asserted-by": "publisher",
"first-page": "12",
"journal-title": "Virus Res.",
"key": "D4CB00230J/cit7/1",
"volume": "202",
"year": "2015"
},
{
"DOI": "10.1016/j.genrep.2020.100860",
"author": "Gurung",
"doi-asserted-by": "publisher",
"first-page": "100860",
"journal-title": "Gene Rep.",
"key": "D4CB00230J/cit8/1",
"volume": "21",
"year": "2020"
},
{
"DOI": "10.1038/s41598-020-61432-1",
"author": "Jang",
"doi-asserted-by": "publisher",
"first-page": "4481",
"journal-title": "Sci. Rep.",
"key": "D4CB00230J/cit9/1",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1038/s41467-021-25166-6",
"author": "Newman",
"doi-asserted-by": "publisher",
"first-page": "1",
"issue": "1",
"journal-title": "Nat. Commun.",
"key": "D4CB00230J/cit10/1",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1016/j.bpj.2020.11.2276",
"author": "Mickolajczyk",
"doi-asserted-by": "publisher",
"first-page": "1020",
"issue": "6",
"journal-title": "Biophys. J.",
"key": "D4CB00230J/cit11/1",
"volume": "120",
"year": "2021"
},
{
"DOI": "10.1074/jbc.c300328200",
"author": "Tanner",
"doi-asserted-by": "publisher",
"first-page": "39578",
"issue": "41",
"journal-title": "J. Biol. Chem.",
"key": "D4CB00230J/cit12/1",
"volume": "278",
"year": "2003"
},
{
"DOI": "10.1128/jvi.78.11.5619-5632.2004",
"author": "Ivanov",
"doi-asserted-by": "publisher",
"first-page": "5619",
"issue": "11",
"journal-title": "J. Virol.",
"key": "D4CB00230J/cit13/1",
"volume": "78",
"year": "2004"
},
{
"DOI": "10.1371/journal.pone.0036521",
"author": "Adedeji",
"doi-asserted-by": "publisher",
"first-page": "e36521",
"issue": "5",
"journal-title": "PLoS One",
"key": "D4CB00230J/cit14/1",
"volume": "7",
"year": "2012"
},
{
"DOI": "10.1073/pnas.2100170118",
"author": "Wilamowski",
"doi-asserted-by": "publisher",
"first-page": "e2100170118",
"issue": "21",
"journal-title": "Proc. Natl. Acad. Sci. U. S. A.",
"key": "D4CB00230J/cit15/1",
"volume": "118",
"year": "2021"
},
{
"DOI": "10.1126/scisignal.abe1202",
"author": "Rosas-Lemus",
"doi-asserted-by": "publisher",
"first-page": "1202",
"issue": "651",
"journal-title": "Sci. Signaling",
"key": "D4CB00230J/cit16/1",
"volume": "13",
"year": "2020"
},
{
"DOI": "10.1038/s41586-022-05185-z",
"author": "Park",
"doi-asserted-by": "publisher",
"first-page": "793",
"issue": "7928",
"journal-title": "Nature",
"key": "D4CB00230J/cit17/1",
"volume": "609",
"year": "2022"
},
{
"DOI": "10.1038/s41467-020-17496-8",
"author": "Viswanathan",
"doi-asserted-by": "publisher",
"first-page": "1",
"issue": "1",
"journal-title": "Nat. Commun.",
"key": "D4CB00230J/cit18/1",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1038/s41392-022-00884-5",
"author": "Yan",
"doi-asserted-by": "publisher",
"first-page": "1",
"issue": "1",
"journal-title": "Signal Transduction Targeted Ther.",
"key": "D4CB00230J/cit19/1",
"volume": "7",
"year": "2022"
},
{
"DOI": "10.1016/J.CELL.2020.07.033",
"author": "Chen",
"doi-asserted-by": "publisher",
"first-page": "1560",
"issue": "6",
"journal-title": "Cell",
"key": "D4CB00230J/cit20/1",
"volume": "182",
"year": "2020"
},
{
"DOI": "10.1016/j.antiviral.2021.105033",
"author": "Martin",
"doi-asserted-by": "publisher",
"first-page": "105033",
"journal-title": "Antiviral Res.",
"key": "D4CB00230J/cit21/1",
"volume": "188",
"year": "2021"
},
{
"DOI": "10.1128/AAC.00957-12",
"author": "Adedeji",
"doi-asserted-by": "publisher",
"first-page": "4718",
"issue": "9",
"journal-title": "Antimicrob. Agents Chemother.",
"key": "D4CB00230J/cit22/1",
"volume": "56",
"year": "2012"
},
{
"DOI": "10.1016/J.BMCL.2012.04.081",
"author": "Yu",
"doi-asserted-by": "publisher",
"first-page": "4049",
"issue": "12",
"journal-title": "Bioorg. Med. Chem. Lett.",
"key": "D4CB00230J/cit23/1",
"volume": "22",
"year": "2012"
},
{
"DOI": "10.1038/s41467-020-19770-1",
"author": "Yan",
"doi-asserted-by": "publisher",
"first-page": "1",
"issue": "1",
"journal-title": "Nat. Commun.",
"key": "D4CB00230J/cit24/1",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1093/nar/gkz409",
"author": "Jia",
"doi-asserted-by": "publisher",
"first-page": "6538",
"issue": "12",
"journal-title": "Nucleic Acids Res.",
"key": "D4CB00230J/cit25/1",
"volume": "47",
"year": "2019"
},
{
"DOI": "10.4161/RNA.27446",
"author": "Zander",
"doi-asserted-by": "publisher",
"first-page": "45",
"issue": "1",
"journal-title": "RNA Biol.",
"key": "D4CB00230J/cit26/1",
"volume": "11",
"year": "2014"
},
{
"DOI": "10.1093/nar/gkad660",
"author": "Marx",
"doi-asserted-by": "publisher",
"first-page": "9266",
"journal-title": "Nucleic Acids Res.",
"key": "D4CB00230J/cit27/1",
"volume": "17",
"year": "2023"
},
{
"DOI": "10.1016/j.bbrep.2018.10.011",
"author": "Jakob",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Biochem. Biophys. Rep.",
"key": "D4CB00230J/cit28/1",
"volume": "17",
"year": "2019"
},
{
"DOI": "10.1021/acsinfecdis.9b00220",
"author": "Ablenas",
"doi-asserted-by": "publisher",
"first-page": "2118",
"issue": "12",
"journal-title": "ACS Infect. Dis.",
"key": "D4CB00230J/cit29/1",
"volume": "5",
"year": "2019"
},
{
"DOI": "10.1021/ja027007w",
"author": "Chin",
"doi-asserted-by": "publisher",
"first-page": "9026",
"issue": "31",
"journal-title": "J. Am. Chem. Soc.",
"key": "D4CB00230J/cit30/1",
"volume": "124",
"year": "2002"
},
{
"DOI": "10.1021/cr400355w",
"author": "Lang",
"doi-asserted-by": "publisher",
"first-page": "4764",
"issue": "9",
"journal-title": "Chem. Rev.",
"key": "D4CB00230J/cit31/1",
"volume": "114",
"year": "2014"
},
{
"DOI": "10.1126/sciadv.abj4526",
"author": "Perez-Lemus",
"doi-asserted-by": "publisher",
"first-page": "4526",
"issue": "1",
"journal-title": "Sci. Adv.",
"key": "D4CB00230J/cit32/1",
"volume": "8",
"year": "2022"
},
{
"DOI": "10.1042/bcj20210201",
"author": "Zeng",
"doi-asserted-by": "publisher",
"first-page": "2405",
"issue": "13",
"journal-title": "Biochem. J.",
"key": "D4CB00230J/cit33/1",
"volume": "478",
"year": "2021"
},
{
"DOI": "10.1002/cbic.201402154",
"author": "Sun",
"doi-asserted-by": "publisher",
"first-page": "1721",
"issue": "12",
"journal-title": "ChemBioChem",
"key": "D4CB00230J/cit34/1",
"volume": "15",
"year": "2014"
},
{
"DOI": "10.1038/nchem.1919",
"author": "Wang",
"doi-asserted-by": "publisher",
"first-page": "393",
"issue": "5",
"journal-title": "Nat. Chem.",
"key": "D4CB00230J/cit35/1",
"volume": "6",
"year": "2014"
},
{
"DOI": "10.3389/fimmu.2022.912717",
"author": "Herbert",
"doi-asserted-by": "publisher",
"first-page": "2814",
"journal-title": "Front. Immunol.",
"key": "D4CB00230J/cit36/1",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1146/annurev-biochem-060713-035737",
"author": "Chin",
"doi-asserted-by": "publisher",
"first-page": "379",
"journal-title": "Annu. Rev. Biochem.",
"key": "D4CB00230J/cit37/1",
"volume": "83",
"year": "2014"
},
{
"DOI": "10.1016/j.chembiol.2014.09.002",
"author": "McKay",
"doi-asserted-by": "publisher",
"first-page": "1075",
"issue": "9",
"journal-title": "Chem. Biol.",
"key": "D4CB00230J/cit38/1",
"volume": "21",
"year": "2014"
},
{
"DOI": "10.1002/anie.201000401",
"author": "Hoogenboom",
"doi-asserted-by": "publisher",
"first-page": "3415",
"issue": "20",
"journal-title": "Angew. Chem., Int. Ed.",
"key": "D4CB00230J/cit39/1",
"volume": "49",
"year": "2010"
},
{
"DOI": "10.1039/D1SC02775A",
"author": "Berta",
"doi-asserted-by": "publisher",
"first-page": "13492",
"issue": "40",
"journal-title": "Chem. Sci.",
"key": "D4CB00230J/cit40/1",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1016/s0959-440x(00)00204-9",
"author": "Ha",
"doi-asserted-by": "publisher",
"first-page": "287",
"issue": "3",
"journal-title": "Curr. Opin. Struct. Biol.",
"key": "D4CB00230J/cit41/1",
"volume": "11",
"year": "2001"
},
{
"DOI": "10.1021/bi800773f",
"author": "Iqbal",
"doi-asserted-by": "publisher",
"first-page": "7857",
"issue": "30",
"journal-title": "Biochemistry",
"key": "D4CB00230J/cit42/1",
"volume": "47",
"year": "2008"
},
{
"DOI": "10.3390/ijms16046718",
"author": "Shrestha",
"doi-asserted-by": "publisher",
"first-page": "6718",
"issue": "4",
"journal-title": "Int. J. Mol. Sci.",
"key": "D4CB00230J/cit43/1",
"volume": "16",
"year": "2015"
},
{
"DOI": "10.1039/c3cs60201j",
"author": "Hwang",
"doi-asserted-by": "publisher",
"first-page": "1221",
"issue": "4",
"journal-title": "Chem. Soc. Rev.",
"key": "D4CB00230J/cit44/1",
"volume": "43",
"year": "2014"
},
{
"DOI": "10.1017/s1355838200000728",
"author": "Seybert",
"doi-asserted-by": "publisher",
"first-page": "1056",
"issue": "7",
"journal-title": "RNA",
"key": "D4CB00230J/cit45/1",
"volume": "6",
"year": "2000"
},
{
"DOI": "10.1007/s12154-013-0094-5",
"author": "Toseland",
"doi-asserted-by": "publisher",
"first-page": "85",
"issue": "3",
"journal-title": "J. Chem. Biol.",
"key": "D4CB00230J/cit46/1",
"volume": "6",
"year": "2013"
},
{
"DOI": "10.1016/j.tibtech.2005.10.005",
"author": "Chudakov",
"doi-asserted-by": "publisher",
"first-page": "605",
"issue": "12",
"journal-title": "Trends Biotechnol.",
"key": "D4CB00230J/cit47/1",
"volume": "23",
"year": "2005"
},
{
"DOI": "10.1021/ja027007w",
"author": "Chin",
"doi-asserted-by": "publisher",
"first-page": "9026",
"issue": "31",
"journal-title": "J. Am. Chem. Soc.",
"key": "D4CB00230J/cit48/1",
"volume": "124",
"year": "2002"
},
{
"DOI": "10.1038/nmeth.1208",
"author": "Roy",
"doi-asserted-by": "publisher",
"first-page": "507",
"issue": "6",
"journal-title": "Nat. Methods",
"key": "D4CB00230J/cit49/1",
"volume": "5",
"year": "2008"
}
],
"reference-count": 49,
"references-count": 49,
"relation": {},
"resource": {
"primary": {
"URL": "https://xlink.rsc.org/?DOI=D4CB00230J"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Monitoring SARS-CoV-2 Nsp13 helicase binding activity using expanded genetic code techniques",
"type": "journal-article",
"update-policy": "https://doi.org/10.1039/rsc_crossmark_policy"
}