Effects of the cross-talk between PARP12/PARP13 and nonsense mediated RNA decay pathway on RNA stability and replication of SARS-CoV-2
et al., Frontiers in Virology, doi:10.3389/fviro.2025.1691166, Nov 2025
In vitro and mouse study showing that PARP12/PARP13 proteins synergize with the nonsense-mediated RNA decay (NMD) pathway to degrade SARS-CoV-2 RNA and inhibit viral replication.
Lokugamage et al., 20 Nov 2025, peer-reviewed, 6 authors.
Contact: nigarg@utmb.edu.
Effects of the cross-talk between PARP12/PARP13 and nonsense mediated RNA decay pathway on RNA stability and replication of SARS-CoV-2
Frontiers in Virology, doi:10.3389/fviro.2025.1691166
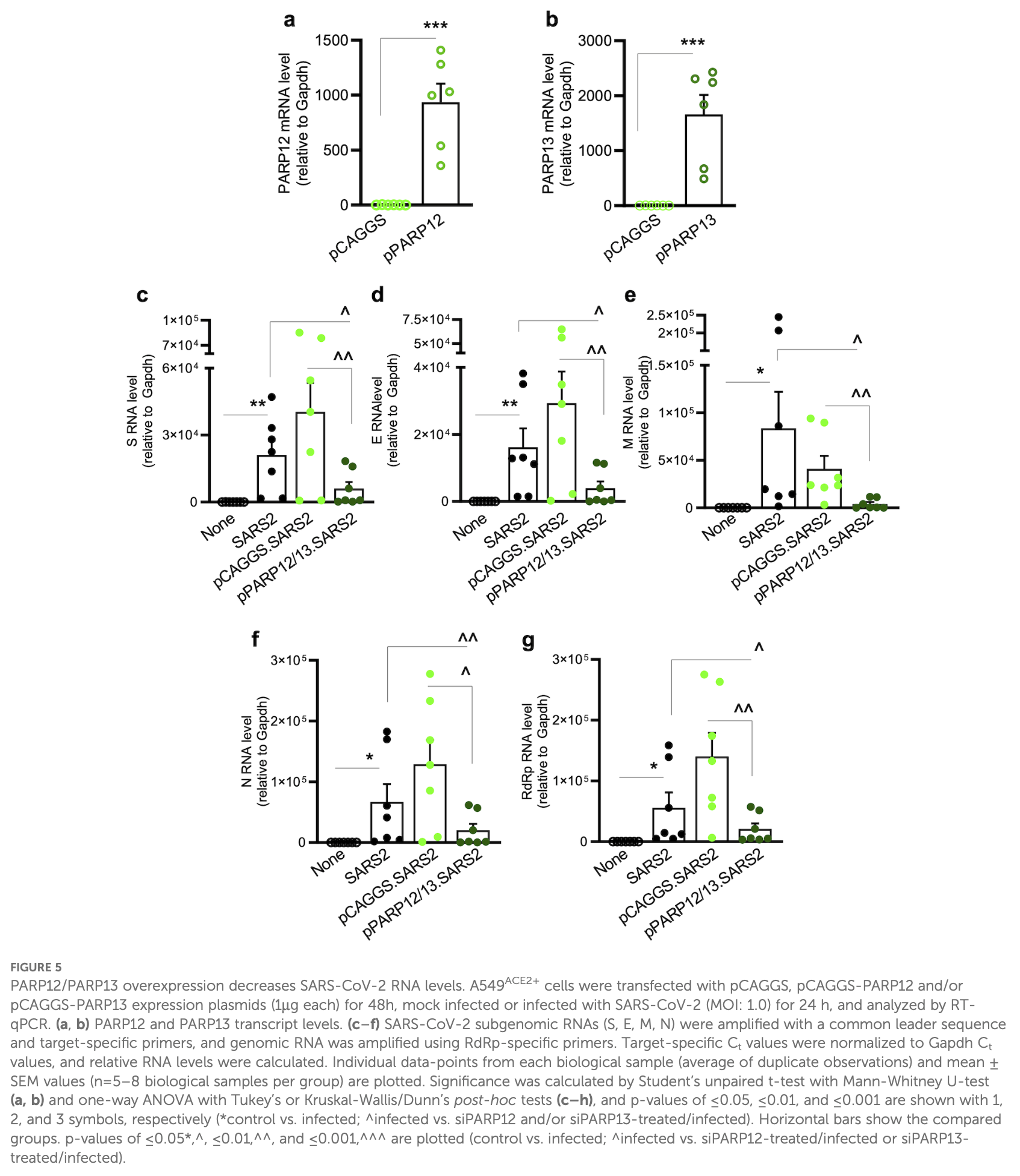
Background: Nonsense-mediated mRNA decay (NMD) pathway recognizes the mRNAs of host and cytoplasmic pathogens harboring aberrant features and targets them for degradation. Poly(ADP-ribose) polymerases (PARPs) superfamily consists of 17 members, among which macrodomain and zinc finger PARPs function as regulators of RNA metabolism and transcription. In this study, we investigated whether crosstalk between NMD and PARPs regulates SARS-CoV-2 RNA stability and viral infection. Methods: Transgenic mice (hACE2 tg ) expressing human angiotensin-converting enzyme 2, and human alveolar epithelial cells (Calu-3 ACE2+ , A549 ACE2+ ), in which the expression of NMD factors and PARPs was modulated by molecular approaches were used for various studies. Results: We found that NMD pathway targets endogenous and exogenous aberrant transcripts in human lung epithelial cells. Upon SARS-CoV-2 infection, the expression of NMD factors, up-framshift 1 and 2 (UPF1/UPF2) was decreased while PARP12 and PARP13 were significantly increased in Calu-3 ACE2+ and A549 ACE2+ cells and lung tissues of hACE2 tg mice. Depletion of PARP12/PARP13 using target-specific (vs. scrambled) siRNAs significantly enhanced the stability of NMD targeted endogenous and exogenous aberrant transcripts and SARS-CoV-2 subgenomic S, E, M, and N mRNAs in A549 ACE2+ cells, like what was noted in siUPF1/ siUPF2-transfected lung epithelial cells. Conversely, overexpression of PARP12/ PARP13 enhanced the NMD-dependent degradation of aberrant transcripts and SARS-CoV-2 subgenomic and genomic RNAs. Further, overexpression of PARP12/ PARP13 had a dose-dependent effect in enhancing the anti-viral NMD activity and suppression of SARS-CoV-2 replication in infected cells.
Conclusion: We conclude that PARP12/PARP13 synergize with NMD pathway to regulate the viral mRNA stability and replication of SARS-CoV-2.
Ethics statement The animal study was approved by Institutional Animal Care and Use Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
Conflict of interest The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement The author(s) declare that no Generative AI was used in the creation of this manuscript. Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Atasheva, Akhrymuk, Frolova, Frolov, New PARP gene with an antialphavirus function, J Virol, doi:10.1128/JVI.00733-12
Atasheva, Frolova, Frolov, Interferon-stimulated poly(ADP-Ribose) polymerases are potent inhibitors of cellular translation and virus replication, J Virol, doi:10.1128/JVI.03443-13
Balistreri, Horvath, Schweingruber, Zund, Mcinerney et al., The host nonsense-mediated mRNA decay pathway restricts Mammalian RNA virus replication, Cell Host Microbe, doi:10.1016/j.chom.2014.08.007
Bick, Carroll, Gao, Goff, Rice et al., Expression of the zinc-finger antiviral protein inhibits alphavirus replication, J Virol, doi:10.1128/jvi.77.21.11555-11562.2003
Blanco-Melo, Nilsson-Payant, Liu, Uhl, Hoagland et al., Imbalanced host response to SARS-coV-2 drives development of COVID-19, Cell, doi:10.1016/j.cell.2020.04.026
Boelz, Neu-Yilik, Gehring, Hentze, Kulozik, A chemiluminescence-based reporter system to monitor nonsense-mediated mRNA decay, Biochem Biophys Res Commun, doi:10.1016/j.bbrc.2006.08.017
Brady, Goel, Johnson, Poly(ADP-ribose) polymerases in hostpathogen interactions, inflammation, and immunity, Microbiol Mol Biol Rev, doi:10.1128/MMBR.00038-18
Busa, Ando, Aigner, Yee, Yeo et al., Transcriptome regulation by PARP13 in basal and antiviral states in human cells, iScience, doi:10.1016/j.isci.2024.109251
Chen, Guo, Lv, Xu, Gao, p72 DEAD box RNA helicase is required for optimal function of the zinc-finger antiviral protein, Proc Natl Acad Sci U.S.A, doi:10.1073/pnas.0712276105
Chiu, Chiu, Yang, Lee, Chiu et al., Inhibition of Japanese encephalitis virus infection by the host zinc-finger antiviral protein, PloS Pathog, doi:10.1371/journal.ppat.1007166
Colombo, Karousis, Bourquin, Bruggmann, Muhlemann, Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6-and SMG7-mediated degradation pathways, RNA, doi:10.1261/rna.059055.116
Ficarelli, Wilson, Galao, Mazzon, Antzin-Anduetza et al., KHNYN is essential for the zinc finger antiviral protein (ZAP) to restrict HIV-1 containing clustered CpG dinucleotides, Elife, doi:10.7554/eLife.46767
Fiorini, Robin, Kanaan, Borowiak, Croquette et al., HTLV-1 Tax plugs and freezes UPF1 helicase leading to nonsense-mediated mRNA decay inhibition, Nat Commun, doi:10.1038/s41467-017-02793-6
Fontaine, Leon, Khalid, Tomar, Jimenez-Morales et al., The cellular NMD pathway restricts zika virus infection and is targeted by the viral capsid protein, mBio, doi:10.1128/mBio.02126-18
Gao, Guo, Goff, Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein, Science, doi:10.1126/science.1074276
Garcia, Garcia, Voinnet, Nonsense-mediated decay serves as a general viral restriction mechanism in plants, Cell Host Microbe, doi:10.1016/j.chom.2014.08.001
Ge, Tian, Huang, Li, Li, An integrative drug repositioning framework discovered a potential therapeutic agent targeting COVID-19, Signal Transduction Targeted Ther, doi:10.1038/s41392-021-00568-6
Gordon, Jang, Bouhaddou, Xu, Obernier et al., A SARS-CoV-2 protein interaction map reveals targets for drug repurposing, Nature, doi:10.1038/s41586-020-2286-9
Grunewald, Shaban, Mackin, Fehr, Perlman, Murine coronavirus infection activates the aryl hydrocarbon receptor in an indoleamine 2,3-dioxygenaseindependent manner, contributing to cytokine modulation and proviral TCDDinducible-PARP expression, J Virol, doi:10.1128/JVI.01743-19
Hayakawa, Shiratori, Yamato, Kameyama, Kitatsuji et al., ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses, Nat Immunol, doi:10.1038/ni.1963
Heer, Sanderson, Voth, Alhammad, Schmidt et al., Coronavirus infection and PARP expression dysregulate the NAD metabolome: An actionable component of innate immunity, J Biol Chem, doi:10.1074/jbc.RA120.015138
Higashi, Maejima, Lee, Yamazaki, Hottiger et al., A study into the ADP-ribosylome of IFN-gamma-stimulated THP-1 human macrophage-like cells identifies ARTD8/PARP14 and ARTD9/PARP9 ADPribosylation, J Proteome Res, doi:10.1021/acs.jproteome.8b00895
Huang, Kraus, The expanding universe of PARP1-mediated molecular and therapeutic mechanisms, Mol Cell, doi:10.1016/j.molcel.2022.02.021
Imamachi, Salam, Suzuki, Akimitsu, A GC-rich sequence feature in the 3' UTR directs UPF1-dependent mRNA decay in mammalian cells, Genome Res, doi:10.1101/gr.206060.116
Iwata, Goettsch, Sharma, Ricchiuto, Goh et al., PARP9 and PARP14 cross-regulate macrophage activation via STAT1 ADP-ribosylation, Nat Commun, doi:10.1038/ncomms12849
Ke, Zhang, Lv, Zeng, Ba, Novel insights into PARPs in gene expression: regulation of RNA metabolism, Cell Mol Life Sci, doi:10.1007/s00018-019-03120-6
Kebaara, Atkin, Long 3'-UTRs target wild-type mRNAs for nonsensemediated mRNA decay in Saccharomyces cerevisiae, Nucleic Acids Res, doi:10.1093/nar/gkp146
Kim, Lee, Yang, Kim, Kim et al., The architecture of SARS-coV-2 transcriptome, Cell, doi:10.1016/j.cell.2020.04.011
Kmiec, Nchioua, Sherrill-Mix, Sturzel, Heusinger et al., CpG frequency in the 5' Third of the env gene determines sensitivity of primary HIV-1 strains to the zinc-finger antiviral protein, mBio, doi:10.1128/mBio.02903-19
Kurosaki, Popp, Maquat, Quality and quantity control of gene expression by nonsense-mediated mRNA decay, Nat Rev Mol Cell Biol, doi:10.1038/s41580-019-0126-2
Li, Zhao, Liu, Li, Quanquin et al., PARP12 suppresses Zika virus infection through PARP-dependent degradation of NS1 and NS3 viral proteins, Sci Signal, doi:10.1126/scisignal.aas9332
Lieberman, Peddu, Xie, Shrestha, Huang et al., In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age, PloS Biol, doi:10.1371/journal.pbio.3000849
Liu, Zhou, Chen, Krug, Battle between influenza A virus and a newly identified antiviral activity of the PARP-containing ZAPL protein, Proc Natl Acad Sci U.S.A, doi:10.1073/pnas.1509745112
Luo, Wang, Gao, Zhu, Liu et al., Molecular mechanism of RNA recognition by zinc-finger antiviral protein, Cell Rep, doi:10.1016/j.celrep.2019.11.116
Mailliot, Vivoli-Vega, Schaffitzel, No-nonsense: insights into the functional interplay of nonsense-mediated mRNA decay factors, Biochem J, doi:10.1042/BCJ20210556
Malgras, Garcia, Jousselin, Bodet, Leveque, The antiviral activities of poly-ADP-ribose polymerases, Viruses, doi:10.3390/v13040582
Mallick, Boehm, Xue, Blackstone, Niels et al., Modulation of UPF1 catalytic activity upon interaction of SARS-CoV-2 Nucleocapsid protein with factors involved in nonsense mediated-mRNA decay, Nucleic Acids Res, doi:10.1093/nar/gkae829
Mao, Nie, Cai, Zhang, Liu et al., Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein, PloS Pathog, doi:10.1371/journal.ppat.1003494
Matyasek, Kovarik, Mutation patterns of human SARS-coV-2 and bat raTG13 coronavirus genomes are strongly biased towards C>U transitions, indicating rapid evolution in their hosts, Genes, doi:10.3390/genes11070761
May, Yuan, Sawicki, Simon, RNA virus evasion of nonsense-mediated decay, PloS Pathog, doi:10.1371/journal.ppat.1007459
Muller, Moller, Bick, Wurr, Becker et al., Inhibition of filovirus replication by the zinc finger antiviral protein, J Virol, doi:10.1128/JVI.01601-06
Nakano, Ando, Yamagishi, Yokoyama, Ishida et al., Viral interference with host mRNA surveillance, the nonsense-mediated mRNA decay (NMD) pathway, through a new function of HTLV-1 Rex: implications for retroviral replication, Microbes Infect, doi:10.1016/j.micinf.2013.03.006
Nuccetelli, Mghezzi-Habellah, Deymier, Roisin, Geŕard-Baraggia et al., The SARS-CoV-2 nucleocapsid protein interferes with the full enzymatic activation of UPF1 and its interaction with UPF2, Nucleic Acids Res, doi:10.1093/nar/gkaf010
Ramage, Kumar, Verschueren, Johnson, Dollen et al., A combined proteomics/genomics approach links hepatitis C virus infection with nonsense-mediated mRNA decay, Mol Cell, doi:10.1016/j.molcel.2014.12.028
Schwerk, Soveg, Ryan, Thomas, Hatfield et al., RNAbinding protein isoforms ZAP-S and ZAP-L have distinct antiviral and immune resolution functions, Nat Immunol, doi:10.1038/s41590-019-0527-6
Shum, Jones, Shao, Chousal, Krause et al., The antagonistic gene paralogs upf3a and upf3b govern nonsense-mediated RNA decay, Cell, doi:10.1016/j.cell.2016.02.046
Thermann, Neu-Yilik, Deters, Frede, Wehr et al., Binary specification of nonsense codons by splicing and cytoplasmic translation, EMBO J, doi:10.1093/emboj/17.12.3484
Todorova, Bock, Chang, PARP13 regulates cellular mRNA posttranscriptionally and functions as a pro-apoptotic factor by destabilizing TRAILR4 transcript, Nat Commun, doi:10.1038/ncomms6362
Tseng, Huang, Newman, Wang, Narayanan et al., Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor, J Virol, doi:10.1128/JVI.01702-06
Wada, Lokugamage, Nakagawa, Narayanan, Makino, Interplay between coronavirus, a cytoplasmic RNA virus, and nonsense-mediated mRNA decay pathway, Proc Natl Acad Sci U.S.A, doi:10.1073/pnas.1811675115
Watson, Barnsley, Toor, Hogan, Winskill et al., Global impact of the first year of COVID-19 vaccination: a mathematical modelling study, Lancet Infect Dis, doi:10.1016/S1473-3099(22)00320-6
Welsby, Hutin, Gueydan, Kruys, Rongvaux et al., PARP12, an interferon-stimulated gene involved in the control of protein translation and inflammation, J Biol Chem, doi:10.1074/jbc.M114.589515
Withers, Beemon, Structural features in the Rous sarcoma virus RNA stability element are necessary for sensing the correct termination codon, Retrovirology, doi:10.1186/1742-4690-7-65
Yoshikawa, Hill, Yoshikawa, Popov, Galindo et al., Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection, PloS One, doi:10.1371/journal.pone.0008729
Younger, Coronavirus 2019: clinical and neuropathological aspects, Curr Opin Rheumatol, doi:10.1097/BOR.0000000000000769
Zhu, Chen, Lv, Wang, Ji et al., Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation, Proc Natl Acad Sci U.S.A, doi:10.1073/pnas.1101676108
Zhu, Wang, Goff, Gao, Translational repression precedes and is required for ZAP-mediated mRNA decay, EMBO J, doi:10.1038/emboj.2012.271
Zimmerlin, Zambidis, Pleiotropic roles of tankyrase/PARP proteins in the establishment and maintenance of human naive pluripotency, Exp Cell Res, doi:10.1016/j.yexcr.2020.111935
DOI record:
{
"DOI": "10.3389/fviro.2025.1691166",
"ISSN": [
"2673-818X"
],
"URL": "http://dx.doi.org/10.3389/fviro.2025.1691166",
"abstract": "<jats:sec>\n <jats:title>Background</jats:title>\n <jats:p>Nonsense-mediated mRNA decay (NMD) pathway recognizes the mRNAs of host and cytoplasmic pathogens harboring aberrant features and targets them for degradation. Poly(ADP-ribose) polymerases (PARPs) superfamily consists of 17 members, among which macrodomain and zinc finger PARPs function as regulators of RNA metabolism and transcription. In this study, we investigated whether crosstalk between NMD and PARPs regulates SARS-CoV-2 RNA stability and viral infection.</jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>Methods</jats:title>\n <jats:p>\n Transgenic mice (hACE2\n <jats:sup>tg</jats:sup>\n ) expressing human angiotensin-converting enzyme 2, and human alveolar epithelial cells (Calu-3\n <jats:sup>ACE2+</jats:sup>\n , A549\n <jats:sup>ACE2+</jats:sup>\n ), in which the expression of NMD factors and PARPs was modulated by molecular approaches were used for various studies.\n </jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>Results</jats:title>\n <jats:p>\n We found that NMD pathway targets endogenous and exogenous aberrant transcripts in human lung epithelial cells. Upon SARS-CoV-2 infection, the expression of NMD factors, up-framshift 1 and 2 (UPF1/UPF2) was decreased while PARP12 and PARP13 were significantly increased in Calu-3\n <jats:sup>ACE2+</jats:sup>\n and A549\n <jats:sup>ACE2+</jats:sup>\n cells and lung tissues of hACE2\n <jats:sup>tg</jats:sup>\n mice. Depletion of PARP12/PARP13 using target-specific (vs. scrambled) siRNAs significantly enhanced the stability of NMD targeted endogenous and exogenous aberrant transcripts and SARS-CoV-2 subgenomic S, E, M, and N mRNAs in A549\n <jats:sup>ACE2+</jats:sup>\n cells, like what was noted in siUPF1/siUPF2-transfected lung epithelial cells. Conversely, overexpression of PARP12/PARP13 enhanced the NMD-dependent degradation of aberrant transcripts and SARS-CoV-2 subgenomic and genomic RNAs. Further, overexpression of PARP12/PARP13 had a dose-dependent effect in enhancing the anti-viral NMD activity and suppression of SARS-CoV-2 replication in infected cells.\n </jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>Conclusion</jats:title>\n <jats:p>We conclude that PARP12/PARP13 synergize with NMD pathway to regulate the viral mRNA stability and replication of SARS-CoV-2.</jats:p>\n </jats:sec>",
"alternative-id": [
"10.3389/fviro.2025.1691166"
],
"article-number": "1691166",
"author": [
{
"affiliation": [],
"family": "Lokugamage",
"given": "Nandadeva",
"sequence": "first"
},
{
"affiliation": [],
"family": "Choudhuri",
"given": "Subhadip",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Rayavara",
"given": "Kempaiah",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Tseng",
"given": "Chien-Te",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Makino",
"given": "Shinji",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Garg",
"given": "Nisha Jain",
"sequence": "additional"
}
],
"container-title": "Frontiers in Virology",
"container-title-short": "Front. Virol.",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"frontiersin.org"
]
},
"created": {
"date-parts": [
[
2025,
11,
20
]
],
"date-time": "2025-11-20T10:31:56Z",
"timestamp": 1763634716000
},
"deposited": {
"date-parts": [
[
2025,
11,
20
]
],
"date-time": "2025-11-20T10:31:57Z",
"timestamp": 1763634717000
},
"indexed": {
"date-parts": [
[
2025,
11,
20
]
],
"date-time": "2025-11-20T10:37:19Z",
"timestamp": 1763635039889,
"version": "3.45.0"
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2025,
11,
20
]
]
},
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
11,
20
]
],
"date-time": "2025-11-20T00:00:00Z",
"timestamp": 1763596800000
}
}
],
"link": [
{
"URL": "https://www.frontiersin.org/articles/10.3389/fviro.2025.1691166/full",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1965",
"original-title": [],
"prefix": "10.3389",
"published": {
"date-parts": [
[
2025,
11,
20
]
]
},
"published-online": {
"date-parts": [
[
2025,
11,
20
]
]
},
"publisher": "Frontiers Media SA",
"reference": [
{
"DOI": "10.1097/BOR.0000000000000769",
"article-title": "Coronavirus 2019: clinical and neuropathological aspects",
"author": "Younger",
"doi-asserted-by": "publisher",
"first-page": "49",
"journal-title": "Curr Opin Rheumatol",
"key": "B1",
"volume": "33",
"year": "2021"
},
{
"DOI": "10.1016/S1473-3099(22)00320-6",
"article-title": "Global impact of the first year of COVID-19 vaccination: a mathematical modelling study",
"author": "Watson",
"doi-asserted-by": "publisher",
"journal-title": "Lancet Infect Dis",
"key": "B2",
"volume": "22",
"year": "2022"
},
{
"DOI": "10.1016/j.cell.2020.04.011",
"article-title": "The architecture of SARS-coV-2 transcriptome",
"author": "Kim",
"doi-asserted-by": "publisher",
"first-page": "914",
"journal-title": "Cell",
"key": "B3",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1016/j.chom.2014.08.007",
"article-title": "The host nonsense-mediated mRNA decay pathway restricts Mammalian RNA virus replication",
"author": "Balistreri",
"doi-asserted-by": "publisher",
"journal-title": "Cell Host Microbe",
"key": "B4",
"volume": "16",
"year": "2014"
},
{
"DOI": "10.1016/j.chom.2014.08.001",
"article-title": "Nonsense-mediated decay serves as a general viral restriction mechanism in plants",
"author": "Garcia",
"doi-asserted-by": "publisher",
"first-page": "391",
"journal-title": "Cell Host Microbe",
"key": "B5",
"volume": "16",
"year": "2014"
},
{
"DOI": "10.1073/pnas.1811675115",
"article-title": "Interplay between coronavirus, a cytoplasmic RNA virus, and nonsense-mediated mRNA decay pathway",
"author": "Wada",
"doi-asserted-by": "publisher",
"journal-title": "Proc Natl Acad Sci U.S.A",
"key": "B6",
"volume": "115",
"year": "2018"
},
{
"DOI": "10.1007/s00018-019-03120-6",
"article-title": "Novel insights into PARPs in gene expression: regulation of RNA metabolism",
"author": "Ke",
"doi-asserted-by": "publisher",
"journal-title": "Cell Mol Life Sci",
"key": "B7",
"volume": "76",
"year": "2019"
},
{
"DOI": "10.1038/s41590-019-0527-6",
"article-title": "RNA-binding protein isoforms ZAP-S and ZAP-L have distinct antiviral and immune resolution functions",
"author": "Schwerk",
"doi-asserted-by": "publisher",
"journal-title": "Nat Immunol",
"key": "B8",
"volume": "20",
"year": "2019"
},
{
"DOI": "10.1038/emboj.2012.271",
"article-title": "Translational repression precedes and is required for ZAP-mediated mRNA decay",
"author": "Zhu",
"doi-asserted-by": "publisher",
"journal-title": "EMBO J",
"key": "B9",
"volume": "31",
"year": "2012"
},
{
"DOI": "10.3390/v13040582",
"article-title": "The antiviral activities of poly-ADP-ribose polymerases",
"author": "Malgras",
"doi-asserted-by": "publisher",
"journal-title": "Viruses",
"key": "B10",
"volume": "13",
"year": "2021"
},
{
"DOI": "10.1371/journal.pone.0008729",
"article-title": "Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection",
"author": "Yoshikawa",
"doi-asserted-by": "publisher",
"first-page": "e8729",
"journal-title": "PloS One",
"key": "B11",
"volume": "5",
"year": "2010"
},
{
"DOI": "10.1128/JVI.01702-06",
"article-title": "Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor",
"author": "Tseng",
"doi-asserted-by": "publisher",
"journal-title": "J Virol",
"key": "B12",
"volume": "81",
"year": "2007"
},
{
"DOI": "10.1093/emboj/17.12.3484",
"article-title": "Binary specification of nonsense codons by splicing and cytoplasmic translation",
"author": "Thermann",
"doi-asserted-by": "publisher",
"journal-title": "EMBO J",
"key": "B13",
"volume": "17",
"year": "1998"
},
{
"DOI": "10.1016/j.bbrc.2006.08.017",
"article-title": "A chemiluminescence-based reporter system to monitor nonsense-mediated mRNA decay",
"author": "Boelz",
"doi-asserted-by": "publisher",
"journal-title": "Biochem Biophys Res Commun",
"key": "B14",
"volume": "349",
"year": "2006"
},
{
"DOI": "10.1016/j.cell.2016.02.046",
"article-title": "The antagonistic gene paralogs upf3a and upf3b govern nonsense-mediated RNA decay",
"author": "Shum",
"doi-asserted-by": "publisher",
"journal-title": "Cell",
"key": "B15",
"volume": "165",
"year": "2016"
},
{
"DOI": "10.1261/rna.059055.116",
"article-title": "Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways",
"author": "Colombo",
"doi-asserted-by": "publisher",
"first-page": "189",
"journal-title": "RNA",
"key": "B16",
"volume": "23",
"year": "2017"
},
{
"DOI": "10.1038/ncomms6362",
"article-title": "PARP13 regulates cellular mRNA post-transcriptionally and functions as a pro-apoptotic factor by destabilizing TRAILR4 transcript",
"author": "Todorova",
"doi-asserted-by": "publisher",
"first-page": "5362",
"journal-title": "Nat Commun",
"key": "B17",
"volume": "5",
"year": "2014"
},
{
"DOI": "10.1016/j.isci.2024.109251",
"article-title": "Transcriptome regulation by PARP13 in basal and antiviral states in human cells",
"author": "Busa",
"doi-asserted-by": "publisher",
"journal-title": "iScience",
"key": "B18",
"volume": "27",
"year": "2024"
},
{
"DOI": "10.1042/BCJ20210556",
"article-title": "No-nonsense: insights into the functional interplay of nonsense-mediated mRNA decay factors",
"author": "Mailliot",
"doi-asserted-by": "publisher",
"journal-title": "Biochem J",
"key": "B19",
"volume": "479",
"year": "2022"
},
{
"DOI": "10.1038/s41580-019-0126-2",
"article-title": "Quality and quantity control of gene expression by nonsense-mediated mRNA decay",
"author": "Kurosaki",
"doi-asserted-by": "publisher",
"journal-title": "Nat Rev Mol Cell Biol",
"key": "B20",
"volume": "20",
"year": "2019"
},
{
"DOI": "10.1093/nar/gkp146",
"article-title": "Long 3'-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae",
"author": "Kebaara",
"doi-asserted-by": "publisher",
"journal-title": "Nucleic Acids Res",
"key": "B21",
"volume": "37",
"year": "2009"
},
{
"DOI": "10.1101/gr.206060.116",
"article-title": "A GC-rich sequence feature in the 3' UTR directs UPF1-dependent mRNA decay in mammalian cells",
"author": "Imamachi",
"doi-asserted-by": "publisher",
"journal-title": "Genome Res",
"key": "B22",
"volume": "27",
"year": "2017"
},
{
"DOI": "10.1128/mBio.02126-18",
"article-title": "The cellular NMD pathway restricts zika virus infection and is targeted by the viral capsid protein",
"author": "Fontaine",
"doi-asserted-by": "publisher",
"journal-title": "mBio",
"key": "B23",
"volume": "9",
"year": "2018"
},
{
"DOI": "10.1016/j.molcel.2014.12.028",
"article-title": "A combined proteomics/genomics approach links hepatitis C virus infection with nonsense-mediated mRNA decay",
"author": "Ramage",
"doi-asserted-by": "publisher",
"journal-title": "Mol Cell",
"key": "B24",
"volume": "57",
"year": "2015"
},
{
"DOI": "10.1186/1742-4690-7-65",
"article-title": "Structural features in the Rous sarcoma virus RNA stability element are necessary for sensing the correct termination codon",
"author": "Withers",
"doi-asserted-by": "publisher",
"journal-title": "Retrovirology",
"key": "B25",
"volume": "7",
"year": "2010"
},
{
"DOI": "10.1371/journal.ppat.1007459",
"article-title": "RNA virus evasion of nonsense-mediated decay",
"author": "May",
"doi-asserted-by": "publisher",
"first-page": "e1007459",
"journal-title": "PloS Pathog",
"key": "B26",
"volume": "14",
"year": "2018"
},
{
"DOI": "10.1038/s41467-017-02793-6",
"article-title": "HTLV-1 Tax plugs and freezes UPF1 helicase leading to nonsense-mediated mRNA decay inhibition",
"author": "Fiorini",
"doi-asserted-by": "publisher",
"first-page": "431",
"journal-title": "Nat Commun",
"key": "B27",
"volume": "9",
"year": "2018"
},
{
"DOI": "10.1016/j.micinf.2013.03.006",
"article-title": "Viral interference with host mRNA surveillance, the nonsense-mediated mRNA decay (NMD) pathway, through a new function of HTLV-1 Rex: implications for retroviral replication",
"author": "Nakano",
"doi-asserted-by": "publisher",
"first-page": "491",
"journal-title": "Microbes Infect",
"key": "B28",
"volume": "15",
"year": "2013"
},
{
"DOI": "10.1038/s41586-020-2286-9",
"article-title": "A SARS-CoV-2 protein interaction map reveals targets for drug repurposing",
"author": "Gordon",
"doi-asserted-by": "publisher",
"journal-title": "Nature",
"key": "B29",
"volume": "583",
"year": "2020"
},
{
"DOI": "10.1093/nar/gkaf010",
"article-title": "The SARS-CoV-2 nucleocapsid protein interferes with the full enzymatic activation of UPF1 and its interaction with UPF2",
"author": "Nuccetelli",
"doi-asserted-by": "publisher",
"journal-title": "Nucleic Acids Res",
"key": "B30",
"volume": "53",
"year": "2025"
},
{
"DOI": "10.1093/nar/gkae829",
"article-title": "Modulation of UPF1 catalytic activity upon interaction of SARS-CoV-2 Nucleocapsid protein with factors involved in nonsense mediated-mRNA decay",
"author": "Mallick",
"doi-asserted-by": "publisher",
"journal-title": "Nucleic Acids Res",
"key": "B31",
"volume": "52",
"year": "2024"
},
{
"DOI": "10.1016/j.molcel.2022.02.021",
"article-title": "The expanding universe of PARP1-mediated molecular and therapeutic mechanisms",
"author": "Huang",
"doi-asserted-by": "publisher",
"journal-title": "Mol Cell",
"key": "B32",
"volume": "82",
"year": "2022"
},
{
"DOI": "10.1038/ncomms12849",
"article-title": "PARP9 and PARP14 cross-regulate macrophage activation via STAT1 ADP-ribosylation",
"author": "Iwata",
"doi-asserted-by": "publisher",
"journal-title": "Nat Commun",
"key": "B33",
"volume": "7",
"year": "2016"
},
{
"DOI": "10.1021/acs.jproteome.8b00895",
"article-title": "A study into the ADP-ribosylome of IFN-gamma-stimulated THP-1 human macrophage-like cells identifies ARTD8/PARP14 and ARTD9/PARP9 ADP-ribosylation",
"author": "Higashi",
"doi-asserted-by": "publisher",
"journal-title": "J Proteome Res",
"key": "B34",
"volume": "18",
"year": "2019"
},
{
"DOI": "10.1016/j.yexcr.2020.111935",
"article-title": "Pleiotropic roles of tankyrase/PARP proteins in the establishment and maintenance of human naive pluripotency",
"author": "Zimmerlin",
"doi-asserted-by": "publisher",
"journal-title": "Exp Cell Res",
"key": "B35",
"volume": "390",
"year": "2020"
},
{
"DOI": "10.1128/MMBR.00038-18",
"article-title": "Poly(ADP-ribose) polymerases in host-pathogen interactions, inflammation, and immunity",
"author": "Brady",
"doi-asserted-by": "publisher",
"journal-title": "Microbiol Mol Biol Rev",
"key": "B36",
"volume": "83",
"year": "2019"
},
{
"DOI": "10.1126/science.1074276",
"article-title": "Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein",
"author": "Gao",
"doi-asserted-by": "publisher",
"journal-title": "Science",
"key": "B37",
"volume": "297",
"year": "2002"
},
{
"DOI": "10.1128/jvi.77.21.11555-11562.2003",
"article-title": "Expression of the zinc-finger antiviral protein inhibits alphavirus replication",
"author": "Bick",
"doi-asserted-by": "publisher",
"journal-title": "J Virol",
"key": "B38",
"volume": "77",
"year": "2003"
},
{
"DOI": "10.1128/JVI.01601-06",
"article-title": "Inhibition of filovirus replication by the zinc finger antiviral protein",
"author": "Muller",
"doi-asserted-by": "publisher",
"journal-title": "J Virol",
"key": "B39",
"volume": "81",
"year": "2007"
},
{
"DOI": "10.1128/JVI.00733-12",
"article-title": "New PARP gene with an anti-alphavirus function",
"author": "Atasheva",
"doi-asserted-by": "publisher",
"journal-title": "J Virol",
"key": "B40",
"volume": "86",
"year": "2012"
},
{
"DOI": "10.1371/journal.ppat.1007166",
"article-title": "Inhibition of Japanese encephalitis virus infection by the host zinc-finger antiviral protein",
"author": "Chiu",
"doi-asserted-by": "publisher",
"first-page": "e1007166",
"journal-title": "PloS Pathog",
"key": "B41",
"volume": "14",
"year": "2018"
},
{
"DOI": "10.1128/JVI.01743-19",
"article-title": "Murine coronavirus infection activates the aryl hydrocarbon receptor in an indoleamine 2,3-dioxygenase-independent manner, contributing to cytokine modulation and proviral TCDD-inducible-PARP expression",
"author": "Grunewald",
"doi-asserted-by": "publisher",
"journal-title": "J Virol",
"key": "B42",
"volume": "94",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.04.026",
"article-title": "Imbalanced host response to SARS-coV-2 drives development of COVID-19",
"author": "Blanco-Melo",
"doi-asserted-by": "publisher",
"first-page": "1036",
"journal-title": "Cell",
"key": "B43",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1371/journal.pbio.3000849",
"article-title": "In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age",
"author": "Lieberman",
"doi-asserted-by": "publisher",
"first-page": "e3000849",
"journal-title": "PloS Biol",
"key": "B44",
"volume": "18",
"year": "2020"
},
{
"DOI": "10.1038/s41392-021-00568-6",
"article-title": "An integrative drug repositioning framework discovered a potential therapeutic agent targeting COVID-19",
"author": "Ge",
"doi-asserted-by": "publisher",
"first-page": "165",
"journal-title": "Signal Transduction Targeted Ther",
"key": "B45",
"volume": "6",
"year": "2021"
},
{
"DOI": "10.1038/ni.1963",
"article-title": "ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses",
"author": "Hayakawa",
"doi-asserted-by": "publisher",
"first-page": "37",
"journal-title": "Nat Immunol",
"key": "B46",
"volume": "12",
"year": "2011"
},
{
"DOI": "10.1371/journal.ppat.1003494",
"article-title": "Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein",
"author": "Mao",
"doi-asserted-by": "publisher",
"first-page": "e1003494",
"journal-title": "PloS Pathog",
"key": "B47",
"volume": "9",
"year": "2013"
},
{
"DOI": "10.1073/pnas.1101676108",
"article-title": "Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation",
"author": "Zhu",
"doi-asserted-by": "publisher",
"journal-title": "Proc Natl Acad Sci U.S.A",
"key": "B48",
"volume": "108",
"year": "2011"
},
{
"DOI": "10.1128/mBio.02903-19",
"article-title": "CpG frequency in the 5' Third of the env gene determines sensitivity of primary HIV-1 strains to the zinc-finger antiviral protein",
"author": "Kmiec",
"doi-asserted-by": "publisher",
"journal-title": "mBio",
"key": "B49",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.3390/genes11070761",
"article-title": "Mutation patterns of human SARS-coV-2 and bat raTG13 coronavirus genomes are strongly biased towards C>U transitions, indicating rapid evolution in their hosts",
"author": "Matyasek",
"doi-asserted-by": "publisher",
"first-page": "761",
"journal-title": "Genes (Basel)",
"key": "B50",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1073/pnas.1509745112",
"article-title": "Battle between influenza A virus and a newly identified antiviral activity of the PARP-containing ZAPL protein",
"author": "Liu",
"doi-asserted-by": "publisher",
"journal-title": "Proc Natl Acad Sci U.S.A",
"key": "B51",
"volume": "112",
"year": "2015"
},
{
"DOI": "10.7554/eLife.46767",
"article-title": "KHNYN is essential for the zinc finger antiviral protein (ZAP) to restrict HIV-1 containing clustered CpG dinucleotides",
"author": "Ficarelli",
"doi-asserted-by": "publisher",
"journal-title": "Elife",
"key": "B52",
"volume": "8",
"year": "2019"
},
{
"DOI": "10.1073/pnas.0712276105",
"article-title": "p72 DEAD box RNA helicase is required for optimal function of the zinc-finger antiviral protein",
"author": "Chen",
"doi-asserted-by": "publisher",
"journal-title": "Proc Natl Acad Sci U.S.A",
"key": "B53",
"volume": "105",
"year": "2008"
},
{
"DOI": "10.1016/j.celrep.2019.11.116",
"article-title": "Molecular mechanism of RNA recognition by zinc-finger antiviral protein",
"author": "Luo",
"doi-asserted-by": "publisher",
"first-page": "46",
"journal-title": "Cell Rep",
"key": "B54",
"volume": "30",
"year": "2020"
},
{
"DOI": "10.1128/JVI.03443-13",
"article-title": "Interferon-stimulated poly(ADP-Ribose) polymerases are potent inhibitors of cellular translation and virus replication",
"author": "Atasheva",
"doi-asserted-by": "publisher",
"journal-title": "J Virol",
"key": "B55",
"volume": "88",
"year": "2014"
},
{
"DOI": "10.1074/jbc.M114.589515",
"article-title": "PARP12, an interferon-stimulated gene involved in the control of protein translation and inflammation",
"author": "Welsby",
"doi-asserted-by": "publisher",
"journal-title": "J Biol Chem",
"key": "B56",
"volume": "289",
"year": "2014"
},
{
"DOI": "10.1126/scisignal.aas9332",
"article-title": "PARP12 suppresses Zika virus infection through PARP-dependent degradation of NS1 and NS3 viral proteins",
"author": "Li",
"doi-asserted-by": "publisher",
"journal-title": "Sci Signal",
"key": "B57",
"volume": "11",
"year": "2018"
},
{
"DOI": "10.1074/jbc.RA120.015138",
"article-title": "Coronavirus infection and PARP expression dysregulate the NAD metabolome: An actionable component of innate immunity",
"author": "Heer",
"doi-asserted-by": "publisher",
"journal-title": "J Biol Chem",
"key": "B58",
"volume": "295",
"year": "2020"
}
],
"reference-count": 58,
"references-count": 58,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.frontiersin.org/articles/10.3389/fviro.2025.1691166/full"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Effects of the cross-talk between PARP12/PARP13 and nonsense mediated RNA decay pathway on RNA stability and replication of SARS-CoV-2",
"type": "journal-article",
"update-policy": "https://doi.org/10.3389/crossmark-policy",
"volume": "5"
}