Dissecting the Interaction Domains of SARS-CoV-2 Nucleocapsid Protein and Human RNA Helicase DDX3X and Search for Potential Inhibitors
et al., International Journal of Molecular Sciences, doi:10.3390/ijms27020672, Jan 2026
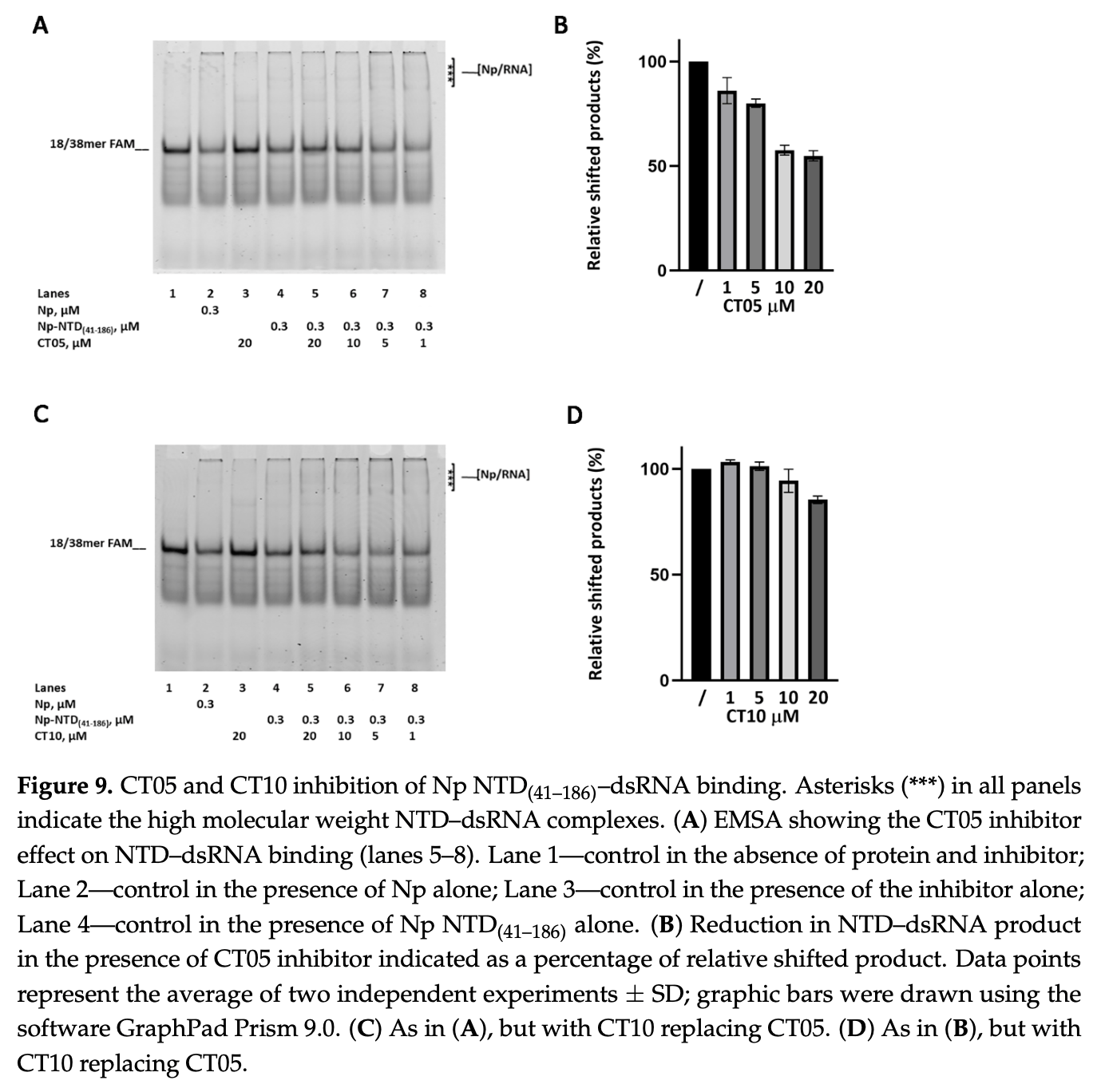
In vitro study showing that two small molecule compounds, CT05 and CT10, inhibit SARS-CoV-2 nucleocapsid protein-RNA binding and may disrupt viral replication.
Lodola et al., 9 Jan 2026, Italy, peer-reviewed, 7 authors.
Contact: massimiliano.secchi@igm.cnr.it (corresponding author), camilla.lodola@igm.cnr.it, mariamichela.pallotta@igm.cnr.it, emmanuele.crespan@igm.cnr.it, paolo.governa@unisi.it.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Dissecting the Interaction Domains of SARS-CoV-2 Nucleocapsid Protein and Human RNA Helicase DDX3X and Search for Potential Inhibitors
International Journal of Molecular Sciences, doi:10.3390/ijms27020672
The SARS-CoV-2 nucleocapsid protein (Np) plays multifunctional roles in the viral life cycle. By interacting with host cellular proteins, Np regulates viral RNA transcription, replication, and immune evasion. It controls genome packaging and counteracts host RNA interference mediated antiviral responses through its RNA binding activity. Previous studies revealed a physical interaction between Np and DDX3X, a human DEAD-box RNA helicase that facilitates the replication of several viruses. This interaction enhances Np affinity for double-stranded RNA and inhibits DDX3X helicase activity. Since Np-RNA binding activity promotes ribonucleoprotein complex formation, targeting this interaction is a promising antiviral strategy. We generated truncated protein variants to define interaction regions between Np and DDX3X. Using AlphaFold modelling, we identified RecA2 as the key DDX3X domain involved in Np binding. Finally, to disrupt Np-RNA complex formation, we screened a small molecule library of putative binders of Np N-terminal region and identified two candidate inhibitors for further development.
Conflicts of Interest: The authors declare no conflicts of interest.
Abbreviations The following abbreviations are used in this manuscript:
References
Abramson, Adler, Dunger, Evans, Green et al., Accurate structure prediction of biomolecular interactions with AlphaFold 3, Nature, doi:10.1038/s41586-024-07487-w
Almeida, Structural basis for the participation of the SARS-CoV-2 nucleocapsid protein in the template switch mechanism and genomic RNA reorganization, J. Biol. Chem, doi:10.1016/j.jbc.2024.107834
Ambike, Cheng, Feuerherd, Velkov, Baldassi et al., Targeting genomic SARS-CoV-2 RNA with siRNAs allows efficient inhibition of viral replication and spread, Nucleic Acids Res, doi:10.1093/nar/gkab1248
Ariumi, Host Cellular RNA Helicases Regulate SARS-CoV-2 Infection, J. Virol, doi:10.1128/jvi.00002-22
Armstrong, Alonso, Garcia-Dorival, Comparative Proteomics and Interactome Analysis of the SARS-CoV-2 Nucleocapsid Protein in Human and Bat Cell Lines, Viruses, doi:10.3390/v16071117
Atkinson, Heaton, Audsley, Kleifeld, Borg, TRIM25 and DEAD-Box RNA Helicase DDX3X Cooperate to Regulate RIG-I-Mediated Antiviral Immunity, Int. J. Mol. Sci, doi:10.3390/ijms22169094
Bai, Cao, Liu, Li, The SARS-CoV-2 Nucleocapsid Protein and Its Role in Viral Structure, Biological Functions, and a Potential Target for Drug or Vaccine Mitigation, Viruses, doi:10.3390/v13061115
Bowden-Reid, Moles, Kelleher, Ahlenstiel, Harnessing antiviral RNAi therapeutics for pandemic viruses: SARS-CoV-2 and HIV, Drug Deliv. Transl. Res, doi:10.1007/s13346-025-01788-x
Brai, Trivisani, Poggialini, Pasqualini, Vagaggini et al., DEAD-Box Helicase DDX3X as a Host Target against Emerging Viruses: New Insights for Medicinal Chemical Approaches, J. Med. Chem, doi:10.1021/acs.jmedchem.2c00755
Carabelli, Peacock, Thorne, Harvey, Hughes, COVID-19
Chen, Farzan, Choe, SARS-CoV-2 spike protein: Structure, viral entry and variants, Nat. Rev. Microbiol, doi:10.1038/s41579-025-01185-8
Ciccosanti, Di Rienzo, Romagnoli, Colavita, Refolo et al., Proteomic analysis identifies the RNA helicase DDX3X as a host target against SARS-CoV-2 infection, Antiviral Res, doi:10.1016/j.antiviral.2021.105064
Cubuk, Alston, Incicco, Singh, Stuchell-Brereton et al., The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA, Nat. Commun, doi:10.1038/s41467-021-21953-3
Di, Dileepan, Ahmed, Liang, Ly, Recombinant SARS-CoV-2 Nucleocapsid Protein: Expression, Purification, and Its Biochemical Characterization and Utility in Serological Assay Development to Assess Immunological Responses to SARS-CoV-2 Infection, Pathogens, doi:10.3390/pathogens10081039
Eiermann, Haneke, Sun, Stoecklin, Ruggieri, Dance with the Devil: Stress Granules and Signaling in Antiviral Responses, Viruses, doi:10.3390/v12090984
Guan, Wang, Fu, Bai, Li et al., Multiple functions of stress granules in viral infection at a glance, Front. Microbiol, doi:10.3389/fmicb.2023.1138864
Hernandez, Martins, Fernandes, Veloso, Lopes et al., Dynamic ensembles of SARS-CoV-2 N-protein reveal head-to-head coiled-coil-driven oligomerization and phase separation, Nucleic Acids Res, doi:10.1093/nar/gkaf502
Huang, Chen, Chen, Huang, Li et al., Molecular characterization of SARS-CoV-2 nucleocapsid protein, Front. Cell. Infect. Microbiol, doi:10.3389/fcimb.2024.1415885
Högbom, Collins, Van Den Berg, Jenvert, Karlberg et al., Crystal structure of conserved domains 1 and 2 of the human DEAD-box helicase DDX3X in complex with the mononucleotide AMP, J. Mol. Biol
Kwon, Choi, Han, A Dual Role of DDX3X in dsRNA-Derived Innate Immune Signaling, Front. Mol. Biosci, doi:10.3389/fmolb.2022.912727
Laughlin, Young, Gonzalez-Gutierrez, Wang, Zlotnick, A narrow ratio of nucleic acid to SARS-CoV-2 N-protein enables phase separation, J. Biol. Chem, doi:10.1016/j.jbc.2024.107831
Li, Hilgenfeld, Whitley, De Clercq, Therapeutic strategies for COVID-19: Progress and lessons learned, Nat Rev Drug Discov, doi:10.1038/s41573-023-00672-y
Li, Yu, Shen, Lai, Liu et al., The RNA-binding proteins regulate innate antiviral immune signaling by modulating pattern recognition receptors, Virol. J, doi:10.1186/s12985-024-02503-x
Liu, Bai, Zhang, Gao, Liu et al., SARS-CoV-2 N Protein Antagonizes Stress Granule Assembly and IFN Production by Interacting with G3BPs to Facilitate Viral Replication, J. Virol, doi:10.1128/jvi.00412-22
Lodola, Secchi, Sinigiani, De Palma, Rossi et al., Interaction of SARS-CoV-2 Nucleocapsid Protein and Human RNA Helicases DDX1 and DDX3X Modulates Their Activities on Double-Stranded RNA, Int. J. Mol. Sci, doi:10.3390/ijms24065784
Ma, Mao, Weng, Teng, Luo et al., DDX3X and virus interactions: Functional diversity and antiviral strategies, Front. Microbiol, doi:10.3389/fmicb.2025.1630068
Min, Huang, Feng, Jia, Sun et al., A New Cellular Interactome of SARS-CoV-2 Nucleocapsid Protein and Its Biological Implications, Mol. Cell. Proteomics, doi:10.1016/j.mcpro.2023.100579
Mo, Liang, Su, Li, Chen et al., DDX3X: Structure, physiologic functions and cancer, Mol. Cancer, doi:10.1186/s12943-021-01325-7
Morse, Sefcikova, Rouzina, Beuning, Williams, Structural domains of SARS-CoV-2 nucleocapsid protein coordinate to compact long nucleic acid substrates, Nucleic Acids Res, doi:10.1093/nar/gkac1179
Mothae, Chiliza, Mvubu, SARS-CoV-2 host-pathogen interactome: Insights into more players during pathogenesis, Virology, doi:10.1016/j.virol.2025.110607
Peacock, Barclay, De Silva, Towers, SARS-CoV-2 variant biology: Immune escape, transmission and fitness, Nat. Rev. Microbiol, doi:10.1038/s41579-022-00841-7
Peng, Du, Lei, Dorje, Qi et al., Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design, EMBO J, doi:10.15252/embj.2020105938
Rabdano, Ruzanova, Vertyachikh, Teplykh, Emelyanova et al., N-protein vaccine is effective against COVID-19: Phase 3, randomized, double-blind, placebo-controlled clinical trial, J. Infect, doi:10.1016/j.jinf.2024.106288
Riva, Garbelli, Casiraghi, Arena, Trivisani et al., Novel alternative ribonucleotide excision repair pathways in human cells by DDX3X and specialized DNA polymerases, Correction in Nucleic Acids Res, doi:10.1093/nar/gkaa948
Riva, Maga, From the magic bullet to the magic target: Exploiting the diverse roles of DDX3X in viral infections and tumorigenesis, Future Med. Chem, doi:10.4155/fmc-2018-0451
Saha, Ghosh Roy, Dwivedi, Tripathi, Kumar et al., Beyond the Pandemic Era: Recent Advances and Efficacy of SARS-CoV-2 Vaccines Against Emerging Variants of Concern, Vaccines, doi:10.3390/vaccines13040424
Samir, Kanneganti, DEAD/H-Box Helicases in Immunity, Inflammation, Cell Differentiation, and Cell Death and Disease, Cells, doi:10.3390/cells11101608
Savastano, Ibáñez De Opakua, Rankovic, Zweckstetter, Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates, Nat. Commun, doi:10.1038/s41467-020-19843-1
Schrödinger, Release, Maestro, Schrödinger
Secchi, Garbelli, Riva, Deidda, Santonicola et al., Synergistic action of human RNaseH2 and the RNA helicase-nuclease DDX3X in processing R-loops, Nucleic Acids Res, doi:10.1093/nar/gkae731
Secchi, Lodola, Garbelli, Bione, Maga, DEAD-Box RNA Helicases DDX3X and DDX5 as Oncogenes or Oncosuppressors: A Network Perspective, Cancers, doi:10.3390/cancers14153820
Tapescu, Cherry, DDX RNA helicases: Key players in cellular homeostasis and innate antiviral immunity, J. Virol, doi:10.1128/jvi.00040-24
Winnard, Jr, Vesuna, Raman, Targeting host DEAD-box RNA helicase DDX3X for treating viral infections, Antiviral Res, doi:10.1016/j.antiviral.2020.104994
Wu, Cheng, Zhou, Sun, Zhang, The SARS-CoV-2 nucleocapsid protein: Its role in the viral life cycle, structure and functions, and use as a potential target in the development of vaccines and diagnostics, Virol. J, doi:10.1186/s12985-023-01968-6
Yang, Johnson, Meliopoulos, Ju, Zhang et al., Interaction between host G3BP and viral nucleocapsid protein regulates SARS-CoV-2 replication and pathogenicity, Cell Rep, doi:10.1016/j.celrep.2024.113965
Zhao, Nguyen, Wu, Li, Hassan et al., Plasticity in structure and assembly of SARS-CoV-2 nucleocapsid protein, PNAS Nexus, doi:10.1093/pnasnexus/pgac049
Zhao, Syed, Khalid, Nguyen, Ciling et al., Assembly of SARS-CoV-2 nucleocapsid protein with nucleic acid, Nucleic Acids Res, doi:10.1093/nar/gkae256
Zhong, Chen, Lu, Luo, Shi et al., Liquid-liquid phase separation of DDX3X: Mechanisms, pathological implications, and therapeutic potential, Int. J. Biol. Macromol, doi:10.1016/j.ijbiomac.2025.144835
DOI record:
{
"DOI": "10.3390/ijms27020672",
"ISSN": [
"1422-0067"
],
"URL": "http://dx.doi.org/10.3390/ijms27020672",
"abstract": "<jats:p>The SARS-CoV-2 nucleocapsid protein (Np) plays multifunctional roles in the viral life cycle. By interacting with host cellular proteins, Np regulates viral RNA transcription, replication, and immune evasion. It controls genome packaging and counteracts host RNA interference mediated antiviral responses through its RNA binding activity. Previous studies revealed a physical interaction between Np and DDX3X, a human DEAD-box RNA helicase that facilitates the replication of several viruses. This interaction enhances Np affinity for double-stranded RNA and inhibits DDX3X helicase activity. Since Np-RNA binding activity promotes ribonucleoprotein complex formation, targeting this interaction is a promising antiviral strategy. We generated truncated protein variants to define interaction regions between Np and DDX3X. Using AlphaFold modelling, we identified RecA2 as the key DDX3X domain involved in Np binding. Finally, to disrupt Np-RNA complex formation, we screened a small molecule library of putative binders of Np N-terminal region and identified two candidate inhibitors for further development.</jats:p>",
"alternative-id": [
"ijms27020672"
],
"author": [
{
"ORCID": "https://orcid.org/0009-0000-2233-0762",
"affiliation": [
{
"name": "Institute of Molecular Genetics IGM-CNR ‘Luigi Luca Cavalli-Sforza’, Via Abbiategrasso 207, I-27100 Pavia, Italy"
}
],
"authenticated-orcid": false,
"family": "Lodola",
"given": "Camilla",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0000-0001-7731-8621",
"affiliation": [
{
"name": "Institute of Molecular Genetics IGM-CNR ‘Luigi Luca Cavalli-Sforza’, Via Abbiategrasso 207, I-27100 Pavia, Italy"
}
],
"authenticated-orcid": false,
"family": "Pallotta",
"given": "Maria Michela",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-9598-2339",
"affiliation": [
{
"name": "Department of Biotechnology, Chemistry and Pharmacy, University of Siena, Via Aldo Moro 2, I-53100 Siena, Italy"
}
],
"authenticated-orcid": false,
"family": "Manetti",
"given": "Fabrizio",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-5976-780X",
"affiliation": [
{
"name": "Department of Biotechnology, Chemistry and Pharmacy, University of Siena, Via Aldo Moro 2, I-53100 Siena, Italy"
},
{
"name": "Department of Food and Drug, University of Parma, Parco Area delle Scienze 27/A, I-43124 Parma, Italy"
}
],
"authenticated-orcid": false,
"family": "Governa",
"given": "Paolo",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Molecular Genetics IGM-CNR ‘Luigi Luca Cavalli-Sforza’, Via Abbiategrasso 207, I-27100 Pavia, Italy"
}
],
"family": "Crespan",
"given": "Emmanuele",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Molecular Genetics IGM-CNR ‘Luigi Luca Cavalli-Sforza’, Via Abbiategrasso 207, I-27100 Pavia, Italy"
}
],
"family": "Maga",
"given": "Giovanni",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-4570-1885",
"affiliation": [
{
"name": "Institute of Molecular Genetics IGM-CNR ‘Luigi Luca Cavalli-Sforza’, Via Abbiategrasso 207, I-27100 Pavia, Italy"
}
],
"authenticated-orcid": false,
"family": "Secchi",
"given": "Massimiliano",
"sequence": "additional"
}
],
"container-title": "International Journal of Molecular Sciences",
"container-title-short": "IJMS",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2026,
1,
9
]
],
"date-time": "2026-01-09T10:35:52Z",
"timestamp": 1767954952000
},
"deposited": {
"date-parts": [
[
2026,
1,
11
]
],
"date-time": "2026-01-11T05:16:47Z",
"timestamp": 1768108607000
},
"indexed": {
"date-parts": [
[
2026,
1,
11
]
],
"date-time": "2026-01-11T07:47:16Z",
"timestamp": 1768117636043,
"version": "3.49.0"
},
"is-referenced-by-count": 0,
"issue": "2",
"issued": {
"date-parts": [
[
2026,
1,
9
]
]
},
"journal-issue": {
"issue": "2",
"published-online": {
"date-parts": [
[
2026,
1
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2026,
1,
9
]
],
"date-time": "2026-01-09T00:00:00Z",
"timestamp": 1767916800000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/1422-0067/27/2/672/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "672",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2026,
1,
9
]
]
},
"published-online": {
"date-parts": [
[
2026,
1,
9
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.1016/j.virol.2025.110607",
"article-title": "SARS-CoV-2 host-pathogen interactome: Insights into more players during pathogenesis",
"author": "Mothae",
"doi-asserted-by": "crossref",
"first-page": "110607",
"journal-title": "Virology",
"key": "ref_1",
"volume": "610",
"year": "2025"
},
{
"DOI": "10.1093/nar/gkac1179",
"article-title": "Structural domains of SARS-CoV-2 nucleocapsid protein coordinate to compact long nucleic acid substrates",
"author": "Morse",
"doi-asserted-by": "crossref",
"first-page": "290",
"journal-title": "Nucleic Acids Res.",
"key": "ref_2",
"volume": "51",
"year": "2023"
},
{
"DOI": "10.1093/nar/gkae256",
"article-title": "Assembly of SARS-CoV-2 nucleocapsid protein with nucleic acid",
"author": "Zhao",
"doi-asserted-by": "crossref",
"first-page": "6647",
"journal-title": "Nucleic Acids Res.",
"key": "ref_3",
"volume": "52",
"year": "2024"
},
{
"DOI": "10.1016/j.jbc.2024.107834",
"article-title": "Structural basis for the participation of the SARS-CoV-2 nucleocapsid protein in the template switch mechanism and genomic RNA reorganization",
"author": "Bezerra",
"doi-asserted-by": "crossref",
"first-page": "107834",
"journal-title": "J. Biol. Chem.",
"key": "ref_4",
"volume": "300",
"year": "2024"
},
{
"DOI": "10.1016/j.jbc.2024.107831",
"article-title": "A narrow ratio of nucleic acid to SARS-CoV-2 N-protein enables phase separation",
"author": "Laughlin",
"doi-asserted-by": "crossref",
"first-page": "107831",
"journal-title": "J. Biol. Chem.",
"key": "ref_5",
"volume": "300",
"year": "2024"
},
{
"DOI": "10.1093/nar/gkaf502",
"article-title": "Dynamic ensembles of SARS-CoV-2 N-protein reveal head-to-head coiled-coil-driven oligomerization and phase separation",
"author": "Hernandez",
"doi-asserted-by": "crossref",
"first-page": "gkaf502",
"journal-title": "Nucleic Acids Res.",
"key": "ref_6",
"volume": "53",
"year": "2025"
},
{
"DOI": "10.3390/v13061115",
"doi-asserted-by": "crossref",
"key": "ref_7",
"unstructured": "Bai, Z., Cao, Y., Liu, W., and Li, J. (2021). The SARS-CoV-2 Nucleocapsid Protein and Its Role in Viral Structure, Biological Functions, and a Potential Target for Drug or Vaccine Mitigation. Viruses, 13."
},
{
"DOI": "10.1186/s12985-023-01968-6",
"article-title": "The SARS-CoV-2 nucleocapsid protein: Its role in the viral life cycle, structure and functions, and use as a potential target in the development of vaccines and diagnostics",
"author": "Wu",
"doi-asserted-by": "crossref",
"first-page": "6",
"journal-title": "Virol. J.",
"key": "ref_8",
"volume": "20",
"year": "2023"
},
{
"DOI": "10.3389/fcimb.2024.1415885",
"doi-asserted-by": "crossref",
"key": "ref_9",
"unstructured": "Huang, Y., Chen, J., Chen, S., Huang, C., Li, B., Li, J., Jin, Z., Zhang, Q., Pan, P., and Du, W. (2024). Molecular characterization of SARS-CoV-2 nucleocapsid protein. Front. Cell. Infect. Microbiol., 14."
},
{
"DOI": "10.1016/j.mcpro.2023.100579",
"article-title": "A New Cellular Interactome of SARS-CoV-2 Nucleocapsid Protein and Its Biological Implications",
"author": "Min",
"doi-asserted-by": "crossref",
"first-page": "100579",
"journal-title": "Mol. Cell. Proteomics",
"key": "ref_10",
"volume": "22",
"year": "2023"
},
{
"DOI": "10.3390/ijms24065784",
"doi-asserted-by": "crossref",
"key": "ref_11",
"unstructured": "Lodola, C., Secchi, M., Sinigiani, V., De Palma, A., Rossi, R., Perico, D., Mauri, P.L., and Maga, G. (2023). Interaction of SARS-CoV-2 Nucleocapsid Protein and Human RNA Helicases DDX1 and DDX3X Modulates Their Activities on Double-Stranded RNA. Int. J. Mol. Sci., 24."
},
{
"DOI": "10.1016/j.antiviral.2021.105064",
"article-title": "Proteomic analysis identifies the RNA helicase DDX3X as a host target against SARS-CoV-2 infection",
"author": "Ciccosanti",
"doi-asserted-by": "crossref",
"first-page": "105064",
"journal-title": "Antiviral Res.",
"key": "ref_12",
"volume": "190",
"year": "2021"
},
{
"DOI": "10.1186/s12943-021-01325-7",
"article-title": "DDX3X: Structure, physiologic functions and cancer",
"author": "Mo",
"doi-asserted-by": "crossref",
"first-page": "38",
"journal-title": "Mol. Cancer",
"key": "ref_13",
"volume": "20",
"year": "2021"
},
{
"DOI": "10.3390/cancers14153820",
"doi-asserted-by": "crossref",
"key": "ref_14",
"unstructured": "Secchi, M., Lodola, C., Garbelli, A., Bione, S., and Maga, G. (2022). DEAD-Box RNA Helicases DDX3X and DDX5 as Oncogenes or Oncosuppressors: A Network Perspective. Cancers, 14."
},
{
"DOI": "10.3389/fmolb.2022.912727",
"doi-asserted-by": "crossref",
"key": "ref_15",
"unstructured": "Kwon, J., Choi, H., and Han, C. (2022). A Dual Role of DDX3X in dsRNA-Derived Innate Immune Signaling. Front. Mol. Biosci., 9."
},
{
"DOI": "10.3390/cells11101608",
"doi-asserted-by": "crossref",
"key": "ref_16",
"unstructured": "Samir, P., and Kanneganti, T.D. (2022). DEAD/H-Box Helicases in Immunity, Inflammation, Cell Differentiation, and Cell Death and Disease. Cells, 11."
},
{
"DOI": "10.1128/jvi.00040-24",
"article-title": "DDX RNA helicases: Key players in cellular homeostasis and innate antiviral immunity",
"author": "Tapescu",
"doi-asserted-by": "crossref",
"first-page": "e0004024",
"journal-title": "J. Virol.",
"key": "ref_17",
"volume": "98",
"year": "2024"
},
{
"DOI": "10.3390/ijms22169094",
"doi-asserted-by": "crossref",
"key": "ref_18",
"unstructured": "Atkinson, S.C., Heaton, S.M., Audsley, M.D., Kleifeld, O., and Borg, N.A. (2021). TRIM25 and DEAD-Box RNA Helicase DDX3X Cooperate to Regulate RIG-I-Mediated Antiviral Immunity. Int. J. Mol. Sci., 22."
},
{
"DOI": "10.4155/fmc-2018-0451",
"article-title": "From the magic bullet to the magic target: Exploiting the diverse roles of DDX3X in viral infections and tumorigenesis",
"author": "Riva",
"doi-asserted-by": "crossref",
"first-page": "1357",
"journal-title": "Future Med. Chem.",
"key": "ref_19",
"volume": "11",
"year": "2019"
},
{
"DOI": "10.1016/j.antiviral.2020.104994",
"article-title": "Targeting host DEAD-box RNA helicase DDX3X for treating viral infections",
"author": "Winnard",
"doi-asserted-by": "crossref",
"first-page": "104994",
"journal-title": "Antiviral Res.",
"key": "ref_20",
"volume": "185",
"year": "2021"
},
{
"DOI": "10.3389/fmicb.2025.1630068",
"doi-asserted-by": "crossref",
"key": "ref_21",
"unstructured": "Ma, S., Mao, Q., Weng, S., Teng, M., Luo, J., and Zhang, K. (2025). DDX3X and virus interactions: Functional diversity and antiviral strategies. Front. Microbiol., 16."
},
{
"DOI": "10.1128/jvi.00002-22",
"article-title": "Host Cellular RNA Helicases Regulate SARS-CoV-2 Infection",
"author": "Ariumi",
"doi-asserted-by": "crossref",
"first-page": "e0000222",
"journal-title": "J. Virol.",
"key": "ref_22",
"volume": "96",
"year": "2022"
},
{
"DOI": "10.3390/v16071117",
"doi-asserted-by": "crossref",
"key": "ref_23",
"unstructured": "Armstrong, S.D., Alonso, C., and Garcia-Dorival, I. (2024). Comparative Proteomics and Interactome Analysis of the SARS-CoV-2 Nucleocapsid Protein in Human and Bat Cell Lines. Viruses, 16."
},
{
"DOI": "10.1016/j.celrep.2024.113965",
"article-title": "Interaction between host G3BP and viral nucleocapsid protein regulates SARS-CoV-2 replication and pathogenicity",
"author": "Yang",
"doi-asserted-by": "crossref",
"first-page": "113965",
"journal-title": "Cell Rep.",
"key": "ref_24",
"volume": "43",
"year": "2024"
},
{
"DOI": "10.15252/embj.2020105938",
"article-title": "Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design",
"author": "Peng",
"doi-asserted-by": "crossref",
"first-page": "e105938",
"journal-title": "EMBO J.",
"key": "ref_25",
"volume": "39",
"year": "2020"
},
{
"DOI": "10.1038/s41467-021-21953-3",
"article-title": "The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA",
"author": "Cubuk",
"doi-asserted-by": "crossref",
"first-page": "1936",
"journal-title": "Nat. Commun.",
"key": "ref_26",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1093/pnasnexus/pgac049",
"article-title": "Plasticity in structure and assembly of SARS-CoV-2 nucleocapsid protein",
"author": "Zhao",
"doi-asserted-by": "crossref",
"first-page": "pgac049",
"journal-title": "PNAS Nexus",
"key": "ref_27",
"volume": "1",
"year": "2022"
},
{
"DOI": "10.1016/j.jmb.2007.06.050",
"article-title": "Crystal structure of conserved domains 1 and 2 of the human DEAD-box helicase DDX3X in complex with the mononucleotide AMP",
"author": "Collins",
"doi-asserted-by": "crossref",
"first-page": "150",
"journal-title": "J. Mol. Biol.",
"key": "ref_28",
"volume": "372",
"year": "2007"
},
{
"article-title": "SARS-CoV-2 variant biology: Immune escape, transmission and fitness",
"author": "Carabelli",
"first-page": "162",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_29",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.1016/j.jinf.2024.106288",
"article-title": "N-protein vaccine is effective against COVID-19: Phase 3, randomized, double-blind, placebo-controlled clinical trial",
"author": "Rabdano",
"doi-asserted-by": "crossref",
"first-page": "106288",
"journal-title": "J. Infect",
"key": "ref_30",
"volume": "89",
"year": "2024"
},
{
"DOI": "10.3390/vaccines13040424",
"doi-asserted-by": "crossref",
"key": "ref_31",
"unstructured": "Saha, A., Ghosh Roy, S., Dwivedi, R., Tripathi, P., Kumar, K., Nambiar, S.M., and Pathak, R. (2025). Beyond the Pandemic Era: Recent Advances and Efficacy of SARS-CoV-2 Vaccines Against Emerging Variants of Concern. Vaccines, 13."
},
{
"DOI": "10.1093/nar/gkab1248",
"article-title": "Targeting genomic SARS-CoV-2 RNA with siRNAs allows efficient inhibition of viral replication and spread",
"author": "Ambike",
"doi-asserted-by": "crossref",
"first-page": "333",
"journal-title": "Nucleic Acids Res.",
"key": "ref_32",
"volume": "50",
"year": "2022"
},
{
"DOI": "10.1007/s13346-025-01788-x",
"article-title": "Harnessing antiviral RNAi therapeutics for pandemic viruses: SARS-CoV-2 and HIV",
"author": "Moles",
"doi-asserted-by": "crossref",
"first-page": "2301",
"journal-title": "Drug Deliv. Transl. Res.",
"key": "ref_33",
"volume": "15",
"year": "2025"
},
{
"DOI": "10.1038/s41579-025-01185-8",
"article-title": "SARS-CoV-2 spike protein: Structure, viral entry and variants",
"author": "Chen",
"doi-asserted-by": "crossref",
"first-page": "455",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_34",
"volume": "23",
"year": "2025"
},
{
"DOI": "10.1038/s41573-023-00672-y",
"article-title": "Therapeutic strategies for COVID-19: Progress and lessons learned",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "449",
"journal-title": "Nat Rev Drug Discov.",
"key": "ref_35",
"volume": "22",
"year": "2023"
},
{
"DOI": "10.1186/s12985-024-02503-x",
"article-title": "The RNA-binding proteins regulate innate antiviral immune signaling by modulating pattern recognition receptors",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "225",
"journal-title": "Virol. J.",
"key": "ref_36",
"volume": "21",
"year": "2024"
},
{
"DOI": "10.3390/pathogens10081039",
"doi-asserted-by": "crossref",
"key": "ref_37",
"unstructured": "Di, D., Dileepan, M., Ahmed, S., Liang, Y., and Ly, H. (2021). Recombinant SARS-CoV-2 Nucleocapsid Protein: Expression, Purification, and Its Biochemical Characterization and Utility in Serological Assay Development to Assess Immunological Responses to SARS-CoV-2 Infection. Pathogens, 10."
},
{
"DOI": "10.1038/s41467-020-19843-1",
"article-title": "Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates",
"author": "Savastano",
"doi-asserted-by": "crossref",
"first-page": "6041",
"journal-title": "Nat. Commun.",
"key": "ref_38",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1128/jvi.00412-22",
"article-title": "SARS-CoV-2 N Protein Antagonizes Stress Granule Assembly and IFN Production by Interacting with G3BPs to Facilitate Viral Replication",
"author": "Liu",
"doi-asserted-by": "crossref",
"first-page": "e0041222",
"journal-title": "J. Virol.",
"key": "ref_39",
"volume": "96",
"year": "2022"
},
{
"DOI": "10.1016/j.ijbiomac.2025.144835",
"article-title": "Liquid-liquid phase separation of DDX3X: Mechanisms, pathological implications, and therapeutic potential",
"author": "Zhong",
"doi-asserted-by": "crossref",
"first-page": "144835",
"journal-title": "Int. J. Biol. Macromol.",
"key": "ref_40",
"volume": "317",
"year": "2025"
},
{
"DOI": "10.3390/v12090984",
"doi-asserted-by": "crossref",
"key": "ref_41",
"unstructured": "Eiermann, N., Haneke, K., Sun, Z., Stoecklin, G., and Ruggieri, A. (2020). Dance with the Devil: Stress Granules and Signaling in Antiviral Responses. Viruses, 12."
},
{
"DOI": "10.3389/fmicb.2023.1138864",
"doi-asserted-by": "crossref",
"key": "ref_42",
"unstructured": "Guan, Y., Wang, Y., Fu, X., Bai, G., Li, X., Mao, J., Yan, Y., and Hu, L. (2023). Multiple functions of stress granules in viral infection at a glance. Front. Microbiol., 14."
},
{
"DOI": "10.1021/acs.jmedchem.2c00755",
"article-title": "DEAD-Box Helicase DDX3X as a Host Target against Emerging Viruses: New Insights for Medicinal Chemical Approaches",
"author": "Brai",
"doi-asserted-by": "crossref",
"first-page": "10195",
"journal-title": "J. Med. Chem.",
"key": "ref_43",
"volume": "65",
"year": "2022"
},
{
"DOI": "10.1093/nar/gkaa948",
"article-title": "Novel alternative ribonucleotide excision repair pathways in human cells by DDX3X and specialized DNA polymerases",
"author": "Riva",
"doi-asserted-by": "crossref",
"first-page": "11551",
"journal-title": "Nucleic Acids Res.",
"key": "ref_44",
"volume": "48",
"year": "2020"
},
{
"DOI": "10.1093/nar/gkae731",
"article-title": "Synergistic action of human RNaseH2 and the RNA helicase-nuclease DDX3X in processing R-loops",
"author": "Secchi",
"doi-asserted-by": "crossref",
"first-page": "11641",
"journal-title": "Nucleic Acids Res.",
"key": "ref_45",
"volume": "52",
"year": "2024"
},
{
"DOI": "10.1038/s41586-024-07487-w",
"article-title": "Accurate structure prediction of biomolecular interactions with AlphaFold 3",
"author": "Abramson",
"doi-asserted-by": "crossref",
"first-page": "493",
"journal-title": "Nature",
"key": "ref_46",
"volume": "630",
"year": "2024"
},
{
"key": "ref_47",
"unstructured": "Schrödinger, L.L. (2021). Release 2021-1: Maestro, Schrödinger, LLC."
}
],
"reference-count": 47,
"references-count": 47,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/1422-0067/27/2/672"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Dissecting the Interaction Domains of SARS-CoV-2 Nucleocapsid Protein and Human RNA Helicase DDX3X and Search for Potential Inhibitors",
"type": "journal-article",
"update-policy": "https://doi.org/10.3390/mdpi_crossmark_policy",
"volume": "27"
}