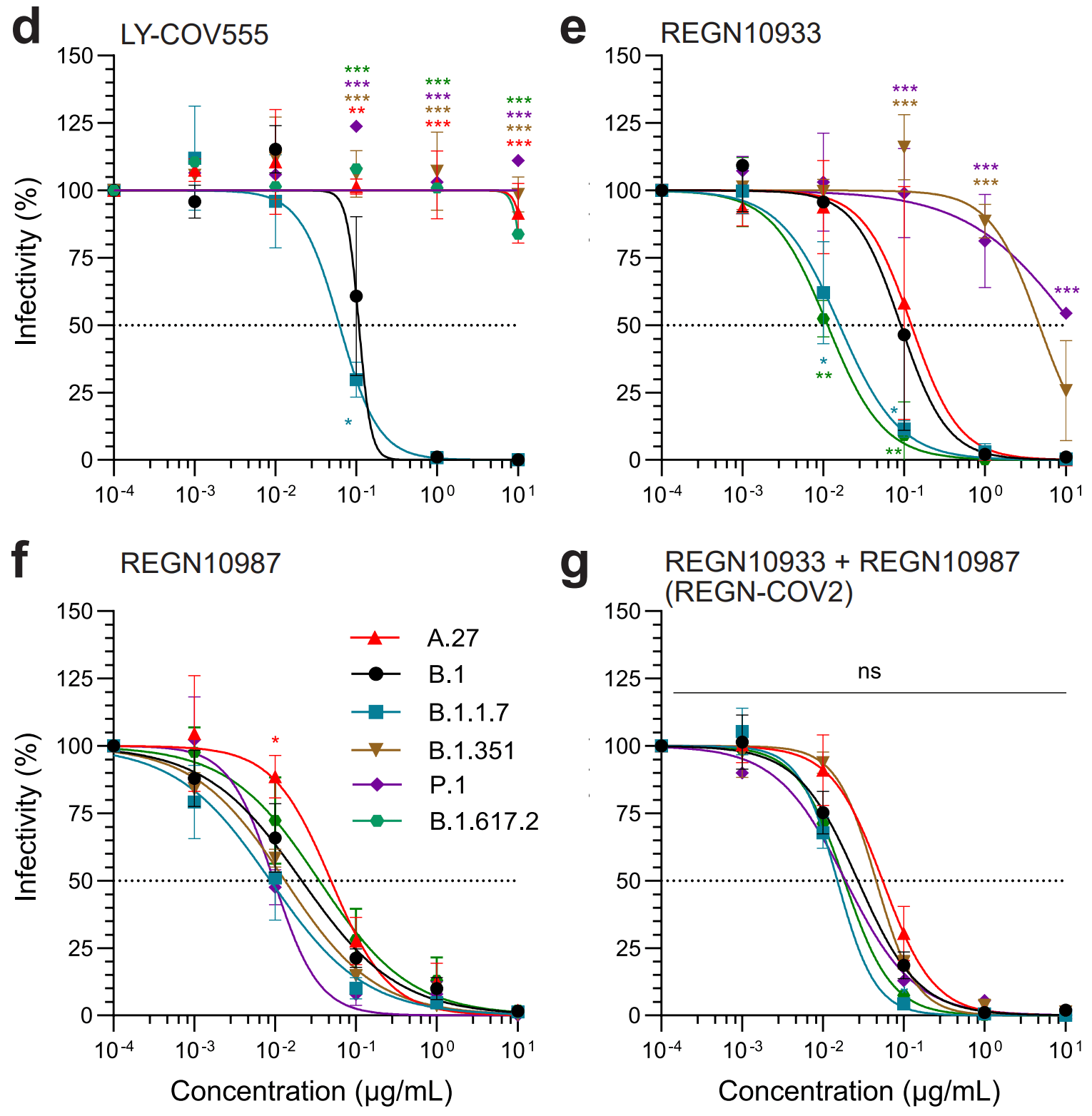
Antibody escape and global spread of SARS-CoV-2 lineage A.27
et al., Research Square, doi:10.21203/rs.3.rs-995033/v1, Nov 2021
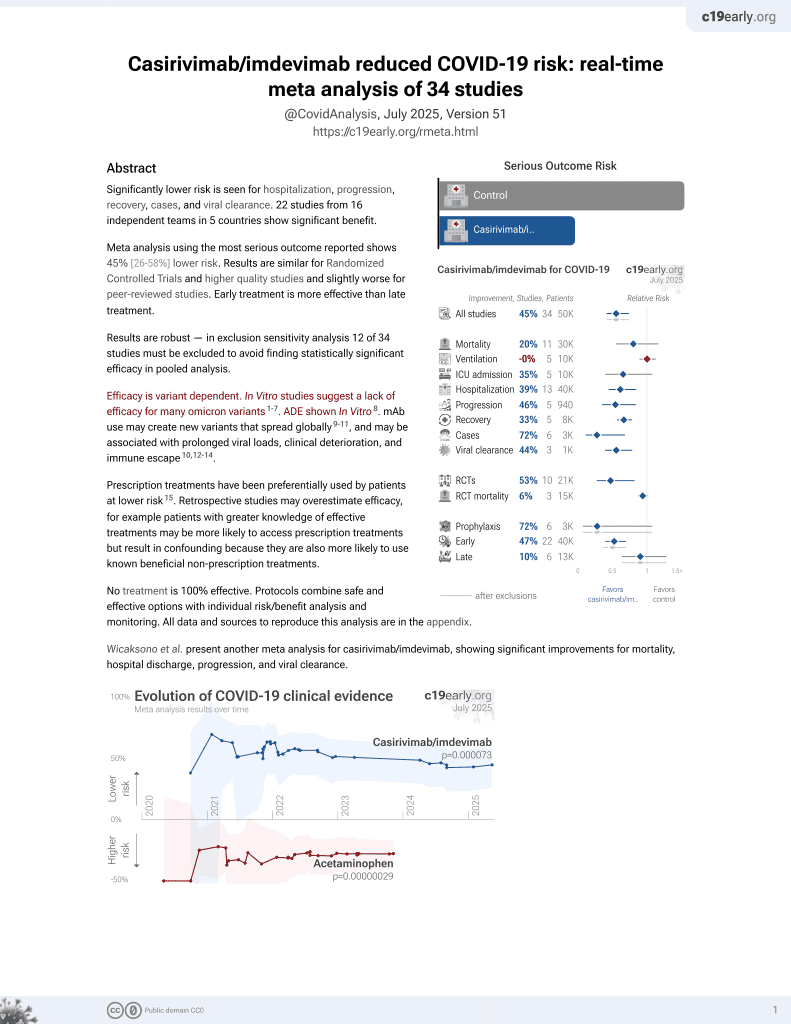
19th treatment shown to reduce risk in
March 2021, now with p = 0.000095 from 34 studies, recognized in 52 countries.
Efficacy is variant dependent.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
Anaysis of antibody escape showing variant A.27 completely escaped neutralization with LY-COV555 and partially with REGN10987. B.1.617.2 escaped these antibodies in a similar manner, suggesting that L452R facilitates the escape. Authors note that B.1.351 and P.1 escaped LY-COV555 and REGN10933, likely facilitated by the E484K mutation, suggesting that L452R and E484K lead to escape from LY-COV555 and to partial resistance to either REGN10987 or REGN10933, respectively.
Efficacy is variant dependent. In Vitro research suggests a lack of efficacy for many omicron variants1-7.
Study covers bamlanivimab/etesevimab and casirivimab/imdevimab.
1.
Liu et al., Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2, bioRxiv, doi:10.1101/2021.12.14.472719.
2.
Sheward et al., Variable loss of antibody potency against SARS-CoV-2 B.1.1.529 (Omicron), bioRxiv, doi:10.1101/2021.12.19.473354.
3.
VanBlargan et al., An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by several therapeutic monoclonal antibodies, bioRxiv, doi:10.1101/2021.12.15.472828.
4.
Tatham et al., Lack of Ronapreve (REGN-CoV; casirivimab and imdevimab) virological efficacy against the SARS-CoV 2 Omicron variant (B.1.1.529) in K18-hACE2 mice, bioRxiv, doi:10.1101/2022.01.23.477397.
5.
Pochtovyi et al., In Vitro Efficacy of Antivirals and Monoclonal Antibodies against SARS-CoV-2 Omicron Lineages XBB.1.9.1, XBB.1.9.3, XBB.1.5, XBB.1.16, XBB.2.4, BQ.1.1.45, CH.1.1, and CL.1, Vaccines, doi:10.3390/vaccines11101533.
Kaleta et al., 2 Nov 2021, preprint, 33 authors.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Antibody escape and global spread of SARS-CoV-2 lineage A.27
doi:10.21203/rs.3.rs-995033/v1
In spring 2021, an increasing number of infections was observed caused by the hitherto rarely described SARS-CoV-2 variant A.27 in south-west Germany. From December 2020 to June 2021 this lineage has been detected in 31 countries. Phylogeographic analyses of A.27 sequences obtained from national and international databases reveal a global spread of this lineage through multiple introductions from its inferred origin in Western Africa. Variant A.27 is characterized by a mutational pattern in the spike gene that includes the L18F, L452R and N501Y spike amino acid substitutions found in various variants of concern but lacks the globally dominant D614G. Neutralization assays demonstrated an escape of A.27 from convalescent and vaccine-elicited antibody-mediated immunity. Moreover, the therapeutic monoclonal antibody Bamlanivimab and partially the REGN-COV2 cocktail failed to block infection by A.27. Our data emphasize the need for continued global monitoring of novel lineages because of the independent evolution of new escape mutations.
Author contributions JF, MH, MP, LK and TK designed the study and contributed to experiment design and data interpretation. MH, JF, SLH, NB, SC, SK and GB collected sequence and associated metadata. SLH, NB and GB performed phylogeographic analyses. JF, LK, MH, SK, SC and AW performed statistical analyses of patient metadata or analysed next-generation sequencing data. AP, AL, MS, NN, JKO, RR, AB, CAK, AO, ESL, VE and EAO provided sequencing data of SARS-CoV-2. TK, LK, JB, DS, SU, SW, GK, PK, MHU and LJ performed experiments and analysed the data. JF, LK, TK, PK, SLH and NB wrote the manuscript. JF, GD and SLH visualized the data. MP, MS and GK were involved in funding acquisition.
Competing interests All authors declare to have no financial or other associations that might pose a potential or actual conflict of interest. Each of these phylogeographic analysis replicates ran for a total of 560 million iterations, respectively, with the Markov chains being sampled every 50,000th iteration, in order to reach an effective sample size (ESS) for all relevant parameters of at least 200, as determined by Tracer 1.7 68 . We used TreeAnnotator to construct maximum clade credibility (MCC) trees for each replicate. The profiles were matched with the R package dplyr, mutations with a frequency below 1% were excluded and the resulting matrix visualized with the R pheatmap package.
Analysis of To identify the lineage-defining mutations, we extracted mutations that were present in 75..
References
Ali, Kasry, Amin, The new SARS-CoV-2 strain shows a stronger binding affinity to ACE2 due to N501Y mutant, Med. drug Discov
Amanat, SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2, Cell
Anoh, SARS-CoV-2 variants of concern, variants of interest and lineage A. 27 are on the rise in Côte d'Ivoire
Ayres, BEAGLE 3: improved performance, scaling, and usability for a highperformance computing library for statistical phylogenetics, Syst. Biol
Baele, Gill, Lemey, Suchard, Hamiltonian Monte Carlo sampling to estimate past population dynamics using the skygrid coalescent model in a Bayesian phylogenetics framework, Wellcome Open Res
Bager, Risk of hospitalisation associated with infection with SARS-CoV-2 lineage B. 1.1. 7 in Denmark: an observational cohort study, Lancet Infect. Dis
Brouwer, Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability, Science
Bugembe, Emergence and spread of a SARS-CoV-2 lineage A variant (A. 23.1) with altered spike protein in Uganda, Nat. Microbiol
Butera, Genomic sequencing of SARS-CoV-2 in Rwanda reveals the importance of incoming travelers on lineage diversity, Nat. Commun
Böhmer, Investigation of a COVID-19 outbreak in Germany resulting from a single travel-associated primary case: a case series, Lancet Infect. Dis
Cerutti, Potent SARS-CoV-2 neutralizing antibodies directed against spike Nterminal domain target a single supersite, Cell Host Microbe
Chen, SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19, N. Engl. J. Med
Chen, Zhou, Chen, Gu, fastp: an ultra-fast all-in-one FASTQ preprocessor, Bioinformatics
Cingolani, A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3, Fly (Austin)
Colson, Spreading of a new SARS-CoV-2 N501Y spike variant in a new lineage, Clin. Microbiol. Infect
Corrales-Aguilar, A novel assay for detecting virus-specific antibodies triggering activation of Fcγ receptors, J. Immunol. Methods
De Maio, Wu, O'reilly, Wilson, New routes to phylogeography: a Bayesian structured coalescent approximation, PLoS Genet
Deng, Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant, Cell
Drummond, Ho, Phillips, Rambaut, Relaxed phylogenetics and dating with confidence, PLoS Biol
Duchene, Temporal signal and the phylodynamic threshold of SARS-CoV-2, Virus Evol
Ferreira, Suchard, Bayesian analysis of elapsed times in continuoustime Markov chains, Can. J. Stat
Flower, Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein, Proc. Natl. Acad. Sci
Geers, SARS-CoV-2 variants of concern partially escape humoral but not Tcell responses in COVID-19 convalescent donors and vaccinees, Sci. Immunol
Gottlieb, Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial, Jama
Greaney, Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies, Cell Host Microbe
Gu, Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy, Science
Hadfield, Nextstrain: real-time tracking of pathogen evolution, Bioinformatics
Hoffmann, SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor, Cell
Hoffmann, SARS-CoV-2 variant B. 1.617 is resistant to Bamlanivimab and evades antibodies induced by infection and vaccination, Cell Rep
Jangra, SARS-CoV-2 spike E484K mutation reduces antibody neutralisation, The Lancet Microbe
Ke, Structures and distributions of SARS-CoV-2 spike proteins on intact virions, Nature
Kern, Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs, Nat. Struct. Mol. Biol
Kolb, Human cytomegalovirus antagonizes activation of Fcγ receptors by distinct and synergizing modes of IgG manipulation, Elife
Korber, Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus, Cell
Krause, SurvNet electronic surveillance system for infectious disease outbreaks, Germany. Emerg. Infect. Dis
Kumar, Bangash, Gruening, Community Research Amid COVID-19 Pandemic: Genomics Analysis of SARS-CoV-2 over Public GALAXY server
Lan, Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor, Nature
Latif, Mullen, Alkuzweny, Tsueng, Cano et al., None
Lauer, The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application, Ann. Intern. Med
Lemey, Accommodating individual travel history and unsampled diversity in Bayesian phylogeographic inference of SARS-CoV-2, Nat. Commun
Lemey, Rambaut, Drummond, Suchard, Bayesian phylogeography finds its roots, PLoS Comput. Biol
Li, Durbin, Fast and accurate short read alignment with Burrows-Wheeler transform, bioinformatics
Li, The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity, Cell
Lustig, Neutralising capacity against Delta (B. 1.617. 2) and other variants of concern following Comirnaty (BNT162b2, BioNTech/Pfizer) vaccination in health care workers, Eurosurveillance
Lynch, Evaluating Markov Chain Monte Carlo Algorithms and Model Fit
Mallm, Local emergence and decline of a SARS-CoV-2 variant with mutations L452R and N501Y in the spike protein, medRxiv
Mccallum, N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2, Cell
Minh, IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era, Mol. Biol. Evol
Minin, Suchard, Counting labeled transitions in continuous-time Markov models of evolution, J. Math. Biol
O'toole, Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool, Virus Evol
Ozono, SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity, Nat. Commun
Pereira, Evolutionary dynamics of the SARS-CoV-2 ORF8 accessory gene, Infect. Genet. Evol
Pirnay, Variant Analysis of SARS-CoV-2 Genomes from Belgian Military Personnel Engaged in Overseas Missions and Operations, Viruses
Premkumar, The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients, Sci. Immunol
Rambaut, A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology, Nat. Microbiol
Rambaut, Drummond, Xie, Baele, Suchard, Posterior summarization in Bayesian phylogenetics using Tracer 1.7, Syst. Biol
Ren, The ORF3a protein of SARS-CoV-2 induces apoptosis in cells, Cell. Mol. Immunol
Sagulenko, Puller, Neher, TreeTime: Maximum-likelihood phylodynamic analysis, Virus Evol
Shu, Mccauley, Gisaid, Global initiative on sharing all influenza data-from vision to reality, Eurosurveillance
Silvas, Contribution of SARS-CoV-2 accessory proteins to viral pathogenicity in K18 hACE2 transgenic mice, J. Virol. JVI
Starr, Greaney, Dingens, Bloom, Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016, Cell Reports Med
Suchard, Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10, Virus Evol
Suryadevara, Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein, Cell
Tennekes, tmap: Thematic Maps in R, J. Stat. Softw
Wang, Structural and functional basis of SARS-CoV-2 entry by using human ACE2, Cell
Weinreich, REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19, N. Engl. J. Med
Wilm, LoFreq: A sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets, Nucleic Acids Res, doi:10.1093/nar/gks918
Winkler, SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function, Nat. Immunol
Wu, The Antigenicity of Epidemic SARS-CoV-2 Variants in the United Kingdom, Front. Immunol
Yan, Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2, Science
Yang, Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods, J. Mol. Evol
Zhang, Structural impact on SARS-CoV-2 spike protein by D614G substitution, Science
Zhang, The SARS-CoV-2 protein ORF3a inhibits fusion of autophagosomes with lysosomes, Cell Discov
Zhou, SARS-CoV-2 spike D614G change enhances replication and transmission, Nature
DOI record:
{
"DOI": "10.21203/rs.3.rs-995033/v1",
"URL": "http://dx.doi.org/10.21203/rs.3.rs-995033/v1",
"abstract": "<jats:title>Abstract</jats:title>\n <jats:p>In spring 2021, an increasing number of infections was observed caused by the hitherto rarely described SARS-CoV-2 variant A.27 in south-west Germany. From December 2020 to June 2021 this lineage has been detected in 31 countries. Phylogeographic analyses of A.27 sequences obtained from national and international databases reveal a global spread of this lineage through multiple introductions from its inferred origin in Western Africa. Variant A.27 is characterized by a mutational pattern in the spike gene that includes the L18F, L452R and N501Y spike amino acid substitutions found in various variants of concern but lacks the globally dominant D614G. Neutralization assays demonstrated an escape of A.27 from convalescent and vaccine-elicited antibody-mediated immunity. Moreover, the therapeutic monoclonal antibody Bamlanivimab and partially the REGN-COV2 cocktail failed to block infection by A.27. Our data emphasize the need for continued global monitoring of novel lineages because of the independent evolution of new escape mutations.</jats:p>",
"accepted": {
"date-parts": [
[
2021,
10,
19
]
]
},
"author": [
{
"affiliation": [
{
"name": "Institute of Virology, Freiburg University Medical Center, Faculty of Medicine, University of Freiburg"
}
],
"family": "Kaleta",
"given": "Tamara",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Institute of Virology, Freiburg University Medical Center, Faculty of Medicine, University of Freiburg"
}
],
"family": "Kern",
"given": "Lisa",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-6354-4943",
"affiliation": [],
"authenticated-orcid": false,
"family": "Hong",
"given": "Samuel",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Methodology and Research Infrastructure, Bioinformatics, Robert Koch Institute"
}
],
"family": "Hölzer",
"given": "Martin",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-0187-559X",
"affiliation": [
{
"name": "Freiburg University Medical Center"
}
],
"authenticated-orcid": false,
"family": "Kochs",
"given": "Georg",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Virology, Freiburg University Medical Center, Faculty of Medicine, University of Freiburg"
}
],
"family": "Beer",
"given": "Julius",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-7528-981X",
"affiliation": [
{
"name": "Institute of Virology"
}
],
"authenticated-orcid": false,
"family": "Schnepf",
"given": "Daniel",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-2972-6855",
"affiliation": [
{
"name": "University Medical Center Freiburg"
}
],
"authenticated-orcid": false,
"family": "Schwemmle",
"given": "Martin",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Microbiology, Immunology and Transplantation, Rega Institute, KU Leuven"
}
],
"family": "Bollen",
"given": "Nena",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-7935-217X",
"affiliation": [
{
"name": "Institute of Virology, Freiburg University Medical Center, Faculty of Medicine, University of Freiburg"
}
],
"authenticated-orcid": false,
"family": "Kolb",
"given": "Philipp",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Virology, Freiburg University Medical Center, Faculty of Medicine, University of Freiburg"
}
],
"family": "Huber",
"given": "Magdalena",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-6605-8163",
"affiliation": [
{
"name": "Institute of Experimental Pharmacology"
}
],
"authenticated-orcid": false,
"family": "Ulferts",
"given": "Svenja",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-8415-6920",
"affiliation": [
{
"name": "Institute of Virology, Freiburg"
}
],
"authenticated-orcid": false,
"family": "Weigang",
"given": "Sebastian",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-0227-4158",
"affiliation": [
{
"name": "Gothenburg Global Biodiversity Centre"
}
],
"authenticated-orcid": false,
"family": "Dudas",
"given": "Gytis",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-0578-8752",
"affiliation": [
{
"name": "Hasso Plattner Institute, Robert Koch Institute"
}
],
"authenticated-orcid": false,
"family": "Wittig",
"given": "Alice",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Virology, Freiburg University Medical Center, Faculty of Medicine, University of Freiburg"
}
],
"family": "Jaki",
"given": "Lena",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institut de Recherche en Santé, de Surveillance Epidémiologique et de Formation (IRESSEF)"
}
],
"family": "Padane",
"given": "Abdou",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Centre de Recherche Médicale et Sanitaire (CERMES)"
}
],
"family": "Lagare",
"given": "Adamou",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Laboratoire de Biologie Moléculaire et d’Immunologie, Département des Sciences Fondamentales, Université de Lomé"
}
],
"family": "Salou",
"given": "Mounerou",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-7131-3691",
"affiliation": [
{
"name": "Northwestern University"
}
],
"authenticated-orcid": false,
"family": "Ozer",
"given": "Egon",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "National Reference Laboratory, Nigeria Centre for Disease Control"
}
],
"family": "Nnaemeka",
"given": "Ndodo",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Virology, Noguchi Memorial Institute for Medical Research, University of Ghana"
}
],
"family": "Odoom",
"given": "John",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Rwanda National Joint Task Force COVID-19, Rwanda Biomedical Centre, Ministry of Health"
}
],
"family": "Rutayisire",
"given": "Robert",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Laboratory of BioInformatics bioMathematics, and bioStatistics (BIMS), Institut Pasteur de Tunis, University of Tunis El Manar"
}
],
"family": "Benkahla",
"given": "Alia",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Laboratoire Central, Centre Hospitalier et Universitaire de Bouaké"
}
],
"family": "Akoua-Koffi",
"given": "Chantal",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Bacteriology and Virology, Souro Sanou University Hospital"
}
],
"family": "Ouedraogo",
"given": "Abdoul-Salam",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-8420-7743",
"affiliation": [
{
"name": "Institut Pasteur"
}
],
"authenticated-orcid": false,
"family": "SIMON-LORIERE",
"given": "Etienne",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Pasteur Institute"
}
],
"family": "Enouf",
"given": "Vincent",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Infectious Disease Epidemiology, Robert Koch Institute"
}
],
"family": "Kröger",
"given": "Stefan",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-4834-0509",
"affiliation": [
{
"name": "Robert Koch Institute"
}
],
"authenticated-orcid": false,
"family": "Calvignac-Spencer",
"given": "Sébastien",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-1915-7732",
"affiliation": [
{
"name": "Rega Institute for Medical Research, KU Leuven"
}
],
"authenticated-orcid": false,
"family": "Baele",
"given": "Guy",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-3596-5964",
"affiliation": [
{
"name": "Institute of Virology, Freiburg University Medical Center, Faculty of Medicine, University of Freiburg"
}
],
"authenticated-orcid": false,
"family": "Panning",
"given": "Marcus",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-1974-212X",
"affiliation": [
{
"name": "Institute of Virology, Freiburg University Medical Center, Faculty of Medicine, University of Freiburg"
}
],
"authenticated-orcid": false,
"family": "Fuchs",
"given": "Jonas",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2021,
11,
2
]
],
"date-time": "2021-11-02T21:39:39Z",
"timestamp": 1635889179000
},
"deposited": {
"date-parts": [
[
2022,
7,
29
]
],
"date-time": "2022-07-29T03:27:53Z",
"timestamp": 1659065273000
},
"group-title": "In Review",
"indexed": {
"date-parts": [
[
2024,
3,
3
]
],
"date-time": "2024-03-03T14:13:59Z",
"timestamp": 1709475239836
},
"institution": [
{
"name": "Research Square"
}
],
"is-referenced-by-count": 1,
"issued": {
"date-parts": [
[
2021,
11,
2
]
]
},
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "unspecified",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
11,
2
]
],
"date-time": "2021-11-02T00:00:00Z",
"timestamp": 1635811200000
}
}
],
"link": [
{
"URL": "https://www.researchsquare.com/article/rs-995033/v1",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.researchsquare.com/article/rs-995033/v1.html",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "8761",
"original-title": [],
"posted": {
"date-parts": [
[
2021,
11,
2
]
]
},
"prefix": "10.21203",
"published": {
"date-parts": [
[
2021,
11,
2
]
]
},
"publisher": "Research Square Platform LLC",
"reference-count": 0,
"references-count": 0,
"relation": {
"is-preprint-of": [
{
"asserted-by": "subject",
"id": "10.1038/s41467-022-28766-y",
"id-type": "doi"
}
]
},
"resource": {
"primary": {
"URL": "https://www.researchsquare.com/article/rs-995033/v1"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"subtype": "preprint",
"title": "Antibody escape and global spread of SARS-CoV-2 lineage A.27",
"type": "posted-content"
}
kaleta