The kinetics of SARS-CoV-2 infection based on a human challenge study
et al., Proceedings of the National Academy of Sciences, doi:10.1073/pnas.2406303121, NCT04865237, Nov 2024
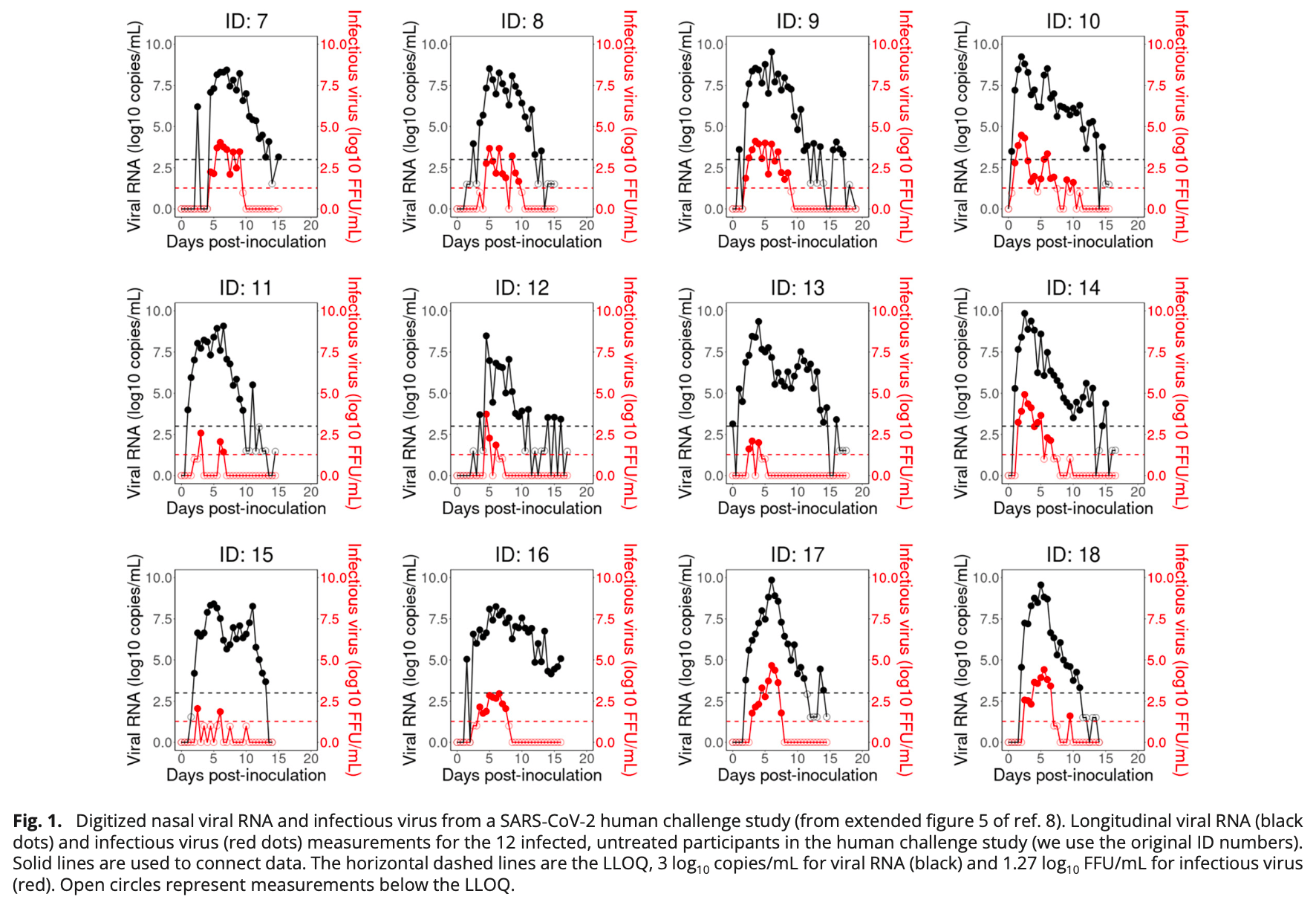
Modeling study of the viral dynamics of SARS-CoV-2 infection using data from a human challenge study. There was a delay of 1-2 days between inoculation and the start of exponential viral growth. During the early stage of infection, viral RNA grew ~1.5 times faster than infectious virus, with a doubling time of ~2 hours for viral RNA and ~3 hours for infectious virus. Productively infected cells have an average lifespan of ~17 hours, spending ~6 hours in the eclipse phase and ~11 hours producing and secreting viruses before dying. Adaptive immune responses, both humoral and cellular, are estimated to be initiated around 7-11 days post-infection. The humoral response leads to a decline in infectious virus by neutralizing antibodies, while the cellular response increases the death rate of infected cells. These contribute to the multi-phasic viral decline observed. Viral rebound experienced by some participants in the later stages is consistent with a decline in the interferon response that puts cells into an antiviral refractory state. As infected cell levels fall, refractory cells transition back to susceptible target cells faster, enabling rebound.
Iyaniwura et al., 7 Nov 2024, peer-reviewed, 6 authors, trial NCT04865237 (history).
Contact: asp@lanl.gov.
Abstract: RESEARCH ARTICLE
SCIENCES
| MEDICAL
PHYSICS
OPEN ACCESS
The kinetics of SARS-CoV-2 infection based on a human
challenge study
Sarafa A. Iyaniwuraa,1
, Ruy M. Ribeiroa
, Carolin Zitzmanna
, Tin Phana
, Ruian Kea
, and Alan S. Perelsona,2
Affiliations are included on p. 9.
Edited by Marcus Feldman, Stanford University, Stanford, CA; received March 28, 2024; accepted October 9, 2024
Studying the early events that occur after viral infection in humans is difficult unless
one intentionally infects volunteers in a human challenge study. Here, we use data
about severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in such a study
in combination with mathematical modeling to gain insights into the relationship
between the amount of virus in the upper respiratory tract and the immune response
it generates. We propose a set of dynamic models of increasing complexity to dissect
the roles of target cell limitation, innate immunity, and adaptive immunity in determining the observed viral kinetics. We introduce an approach for modeling the effect
of humoral immunity that describes a decline in infectious virus after immune activation. We fit our models to viral load and infectious titer data from all the untreated
infected participants in the study simultaneously. We found that a power-law with a
power h < 1 describes the relationship between infectious virus and viral load. Viral
replication at the early stage of infection is rapid, with a doubling time of ~2 h for
viral RNA and ~3 h for infectious virus. We estimate that adaptive immunity is initiated ~7 to 10 d postinfection and appears to contribute to a multiphasic viral decline
experienced by some participants; the viral rebound experienced by other participants
is consistent with a decline in the interferon response. Altogether, we quantified the
kinetics of SARS-CoV-2 infection, shedding light on the early dynamics of the virus
and the potential role of innate and adaptive immunity in promoting viral decline
during infection.
Significance
Studying the early dynamics of
severe acute respiratory
syndrome coronavirus 2 (SARSCoV-2) infection in humans is
difficult. Here, we take advantage
of a detailed dataset from a
human challenge study to fit
dynamic models that include
innate and adaptive immune
responses to longitudinal
changes in both infectious and
total virus. We uncovered a
nonlinear relationship between
total virus and infectious virus.
We found that viral replication,
after a short delay, is rapid, with
a doubling time of ~2 h for viral
RNA and ~3 h for infectious virus.
We also found that innate
immunity wanes as virus is
brought under control, which
together with adaptive immunity,
initiated ~7 to 10 d postinfection,
contributes to a multiphasic viral
decline experienced by some
participants.
SARS-CoV-2 | viral dynamics | human challenge study | infectious disease modeling |
mathematical modeling
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to spread
worldwide, with over 776 million reported cases and 7 million deaths as of August 14,
2024 (1). SARS-CoV-2 is a highly contagious virus that primarily infects cells in the
respiratory tract and lungs and is the causal agent of COVID-19 (2–5) that was first
detected in Wuhan, China, in December 2019 (6).
During acute infection, SARS-CoV-2 viral load, measured by the qPCR in samples
from nose (or throat) swabs, increases rapidly, reaches a peak, and then declines (7–9).
However, qPCR..
DOI record:
{
"DOI": "10.1073/pnas.2406303121",
"ISSN": [
"0027-8424",
"1091-6490"
],
"URL": "http://dx.doi.org/10.1073/pnas.2406303121",
"abstract": "<jats:p>\n Studying the early events that occur after viral infection in humans is difficult unless one intentionally infects volunteers in a human challenge study. Here, we use data about severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in such a study in combination with mathematical modeling to gain insights into the relationship between the amount of virus in the upper respiratory tract and the immune response it generates. We propose a set of dynamic models of increasing complexity to dissect the roles of target cell limitation, innate immunity, and adaptive immunity in determining the observed viral kinetics. We introduce an approach for modeling the effect of humoral immunity that describes a decline in infectious virus after immune activation. We fit our models to viral load and infectious titer data from all the untreated infected participants in the study simultaneously. We found that a power-law with a power\n <jats:italic>h</jats:italic>\n < 1 describes the relationship between infectious virus and viral load. Viral replication at the early stage of infection is rapid, with a doubling time of ~2 h for viral RNA and ~3 h for infectious virus. We estimate that adaptive immunity is initiated ~7 to 10 d postinfection and appears to contribute to a multiphasic viral decline experienced by some participants; the viral rebound experienced by other participants is consistent with a decline in the interferon response. Altogether, we quantified the kinetics of SARS-CoV-2 infection, shedding light on the early dynamics of the virus and the potential role of innate and adaptive immunity in promoting viral decline during infection.\n </jats:p>",
"alternative-id": [
"10.1073/pnas.2406303121"
],
"assertion": [
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2024-03-28"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "2024-10-09"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 3,
"value": "2024-11-07"
}
],
"author": [
{
"ORCID": "http://orcid.org/0000-0002-8854-2335",
"affiliation": [
{
"name": "Theoretical Division, Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, NM 87545"
}
],
"authenticated-orcid": false,
"family": "Iyaniwura",
"given": "Sarafa A.",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0002-3988-8241",
"affiliation": [
{
"name": "Theoretical Division, Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, NM 87545"
}
],
"authenticated-orcid": false,
"family": "Ribeiro",
"given": "Ruy M.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-5188-0042",
"affiliation": [
{
"name": "Theoretical Division, Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, NM 87545"
}
],
"authenticated-orcid": false,
"family": "Zitzmann",
"given": "Carolin",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-9998-8263",
"affiliation": [
{
"name": "Theoretical Division, Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, NM 87545"
}
],
"authenticated-orcid": false,
"family": "Phan",
"given": "Tin",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-5307-8934",
"affiliation": [
{
"name": "Theoretical Division, Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, NM 87545"
}
],
"authenticated-orcid": false,
"family": "Ke",
"given": "Ruian",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-2455-0002",
"affiliation": [
{
"name": "Theoretical Division, Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, NM 87545"
}
],
"authenticated-orcid": false,
"family": "Perelson",
"given": "Alan S.",
"sequence": "additional"
}
],
"container-title": "Proceedings of the National Academy of Sciences",
"container-title-short": "Proc. Natl. Acad. Sci. U.S.A.",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"www.pnas.org"
]
},
"created": {
"date-parts": [
[
2024,
11,
7
]
],
"date-time": "2024-11-07T15:21:24Z",
"timestamp": 1730992884000
},
"deposited": {
"date-parts": [
[
2024,
11,
7
]
],
"date-time": "2024-11-07T15:22:07Z",
"timestamp": 1730992927000
},
"funder": [
{
"DOI": "10.13039/100000002",
"award": [
"U54-HL143541"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100000002",
"id-type": "DOI"
}
],
"name": "HHS | NIH"
},
{
"DOI": "10.13039/100008902",
"award": [
"20200743ER"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100008902",
"id-type": "DOI"
}
],
"name": "DOE | NNSA | Los Alamos National Laboratory"
},
{
"DOI": "10.13039/100008902",
"award": [
"20200695ER"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100008902",
"id-type": "DOI"
}
],
"name": "DOE | NNSA | Los Alamos National Laboratory"
},
{
"DOI": "10.13039/100008902",
"award": [
"20210730ER"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100008902",
"id-type": "DOI"
}
],
"name": "DOE | NNSA | Los Alamos National Laboratory"
}
],
"indexed": {
"date-parts": [
[
2024,
11,
8
]
],
"date-time": "2024-11-08T05:23:57Z",
"timestamp": 1731043437073,
"version": "3.28.0"
},
"is-referenced-by-count": 0,
"issue": "46",
"issued": {
"date-parts": [
[
2024,
11,
7
]
]
},
"journal-issue": {
"issue": "46",
"published-print": {
"date-parts": [
[
2024,
11,
12
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by-nc-nd/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
11,
7
]
],
"date-time": "2024-11-07T00:00:00Z",
"timestamp": 1730937600000
}
}
],
"link": [
{
"URL": "https://pnas.org/doi/pdf/10.1073/pnas.2406303121",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "341",
"original-title": [],
"prefix": "10.1073",
"published": {
"date-parts": [
[
2024,
11,
7
]
]
},
"published-online": {
"date-parts": [
[
2024,
11,
7
]
]
},
"published-print": {
"date-parts": [
[
2024,
11,
12
]
]
},
"publisher": "Proceedings of the National Academy of Sciences",
"reference": [
{
"key": "e_1_3_4_1_2",
"unstructured": "World Health Organization (WHO) Number of COVID-19 deaths reported to WHO. WHO (2024). https://data.who.int/dashboards/covid19/cases?n=c. Accessed 14 March 2024."
},
{
"DOI": "10.1002/jmv.25728",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_2_2"
},
{
"DOI": "10.1128/JVI.01623-05",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_3_2"
},
{
"DOI": "10.1126/science.abc6156",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_4_2"
},
{
"DOI": "10.1038/s41586-020-2196-x",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_5_2"
},
{
"key": "e_1_3_4_6_2",
"unstructured": "World Health Organization (WHO) Timeline: WHO’s COVID-19 response. WHO (2023). https://www.who.int/emergencies/diseases/novel-coronavirus-2019/interactive-timeline. Accessed 14 March 2024."
},
{
"DOI": "10.1016/S2213-2600(22)00226-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_7_2"
},
{
"DOI": "10.1038/s41591-022-01780-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_8_2"
},
{
"DOI": "10.1371/journal.pbio.3001333",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_9_2"
},
{
"DOI": "10.1073/pnas.2111477118",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_10_2"
},
{
"DOI": "10.1038/s41564-022-01105-z",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_11_2"
},
{
"DOI": "10.1371/journal.pcbi.1009997",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_12_2"
},
{
"DOI": "10.7554/eLife.69302",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_13_2"
},
{
"DOI": "10.1101/2022.04.22.22274137",
"doi-asserted-by": "crossref",
"key": "e_1_3_4_14_2",
"unstructured": "J. Carruthers J. Xu T. Finnie I. Hall A within-host model of SARS-CoV-2 infection. medRxiv [Preprint] (2022). https://doi.org/10.1101/2022.04.22.22274137v1 (Accessed 18 January 2024)."
},
{
"DOI": "10.1186/s12916-021-02220-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_15_2"
},
{
"DOI": "10.1371/journal.ppat.1010630",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_16_2"
},
{
"DOI": "10.1126/sciadv.abc7112",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_17_2"
},
{
"DOI": "10.7554/eLife.63537",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_18_2"
},
{
"DOI": "10.1016/j.isci.2022.104448",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_19_2"
},
{
"DOI": "10.1073/pnas.2017962118",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_20_2"
},
{
"DOI": "10.1002/cpt.2160",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_21_2"
},
{
"DOI": "10.1016/j.mbs.2020.108438",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_22_2"
},
{
"DOI": "10.1371/journal.pcbi.1008752",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_23_2"
},
{
"DOI": "10.7554/eLife.75427",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_24_2"
},
{
"DOI": "10.1371/journal.pcbi.1008785",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_25_2"
},
{
"DOI": "10.1093/jac/dkac048",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_26_2"
},
{
"DOI": "10.1101/2023.03.05.23286816",
"doi-asserted-by": "crossref",
"key": "e_1_3_4_27_2",
"unstructured": "G. Lingas Modelling the association between neutralizing antibody levels and SARS-CoV-2 viral dynamics: Implications to define correlates of protection against infection. medRxiv [Preprint] (2023). https://doi.org/10.1101/2023.03.05.23286816."
},
{
"DOI": "10.1371/journal.pcbi.1010721",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_28_2"
},
{
"DOI": "10.3390/v13081635",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_29_2"
},
{
"DOI": "10.1371/journal.pmed.1003660",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_30_2"
},
{
"key": "e_1_3_4_31_2",
"unstructured": "A. Rohatgi WebPlotDigitizer–Copyright 2010–2022 Ankit Rohatgi. Automeris.io (2022). https://apps.automeris.io/wpd/. Accessed 24 February 2023."
},
{
"DOI": "10.1126/sciimmunol.adj9285",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_32_2"
},
{
"DOI": "10.1111/imr.12687",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_33_2"
},
{
"DOI": "10.1126/science.282.5386.103",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_34_2"
},
{
"DOI": "10.1038/nri700",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_35_2"
},
{
"DOI": "10.1006/jtbi.1997.0548",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_36_2"
},
{
"DOI": "10.1002/psp4.12543",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_37_2"
},
{
"DOI": "10.1126/science.1125676",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_38_2"
},
{
"DOI": "10.1371/journal.ppat.1008737",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_39_2"
},
{
"DOI": "10.1128/CMR.14.4.778-809.2001",
"doi-asserted-by": "crossref",
"key": "e_1_3_4_40_2",
"unstructured": "C. E. Samuel Antiviral actions of interferons. Clin. Microbiol. Rev. 14 778–809 (2001)."
},
{
"DOI": "10.1371/journal.ppat.1011680",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_41_2"
},
{
"DOI": "10.1016/S0021-9258(18)33834-1",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_42_2"
},
{
"DOI": "10.1007/978-1-4419-9863-7_946",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_43_2"
},
{
"key": "e_1_3_4_44_2",
"unstructured": "S. A. S. Lixoft Monolix|Lixoft. Lixoft (2015). https://lixoft.com/products/monolix/. Accessed 20 January 2023."
},
{
"DOI": "10.1007/978-1-4757-2917-7_3",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_45_2"
},
{
"DOI": "10.1016/j.cell.2020.05.042",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_46_2"
},
{
"DOI": "10.1099/jgv.0.001453",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_47_2"
},
{
"DOI": "10.1101/2023.05.30.23290747",
"doi-asserted-by": "crossref",
"key": "e_1_3_4_48_2",
"unstructured": "A. S. Perelson R. M. Ribeiro T. Phan An explanation for SARS-CoV-2 rebound after Paxlovid treatment. medRxiv [Preprint] (2023). https://www.medrxiv.org/content/ (Accessed 15 February 2024)."
},
{
"DOI": "10.1016/j.virol.2016.05.019",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_49_2"
},
{
"DOI": "10.1016/S0021-9258(18)45454-3",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_50_2"
},
{
"DOI": "10.7554/eLife.81849",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_51_2"
},
{
"DOI": "10.1371/journal.pcbi.1011437",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_52_2"
},
{
"DOI": "10.1128/JVI.02078-09",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_53_2"
},
{
"DOI": "10.1038/s41586-024-07575-x",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_54_2"
},
{
"DOI": "10.1371/journal.pcbi.1001058",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_55_2"
},
{
"DOI": "10.1002/jmv.25866",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_56_2"
},
{
"DOI": "10.7554/eLife.60122",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_57_2"
},
{
"DOI": "10.1016/j.cell.2021.01.007",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_58_2"
},
{
"DOI": "10.4049/jimmunol.1400844",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_59_2"
},
{
"DOI": "10.1016/j.tim.2017.02.003",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_60_2"
},
{
"DOI": "10.1038/nmicrobiol.2017.78",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_61_2"
},
{
"DOI": "10.1126/science.adg8488",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_62_2"
},
{
"DOI": "10.1084/jem.20210583",
"doi-asserted-by": "crossref",
"key": "e_1_3_4_63_2",
"unstructured": "N. R. Cheemarla Dynamic innate immune response determines susceptibility to SARS-CoV-2 infection and early replication kinetics. J. Exp. Med. 218 e20210583 (2021)."
},
{
"DOI": "10.1038/s43856-022-00195-4",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_64_2"
},
{
"DOI": "10.1126/sciimmunol.abl4509",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_65_2"
},
{
"DOI": "10.1038/s41590-021-01122-w",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_66_2"
},
{
"DOI": "10.1038/s41564-022-01254-1",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_67_2"
},
{
"DOI": "10.7326/M22-2381",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_68_2"
},
{
"DOI": "10.1016/j.lanepe.2021.100164",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_69_2"
},
{
"DOI": "10.3390/v13081642",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_70_2"
},
{
"DOI": "10.7326/M23-1756",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_71_2"
},
{
"DOI": "10.1056/NEJMc2205944",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_72_2"
},
{
"DOI": "10.1016/S1473-3099(23)00063-4",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_73_2"
},
{
"DOI": "10.1016/j.jtbi.2023.111447",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_74_2"
},
{
"DOI": "10.3390/math10173154",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_75_2"
},
{
"DOI": "10.1006/jtbi.2000.1076",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_76_2"
},
{
"DOI": "10.1038/s41598-022-18683-x",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_77_2"
},
{
"DOI": "10.1137/090757009",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_78_2"
}
],
"reference-count": 78,
"references-count": 78,
"relation": {},
"resource": {
"primary": {
"URL": "https://pnas.org/doi/10.1073/pnas.2406303121"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "The kinetics of SARS-CoV-2 infection based on a human challenge study",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1073/pnas.cm10313",
"volume": "121"
}