Prediction of interactions between the SARS-CoV-2 ORF3a protein and small-molecule ligands using the AND-System cognitive platform, graph neural networks, and molecular
et al., Vavilov Journal of Genetics and Breeding, doi:10.18699/vjgb-25-113, Dec 2025
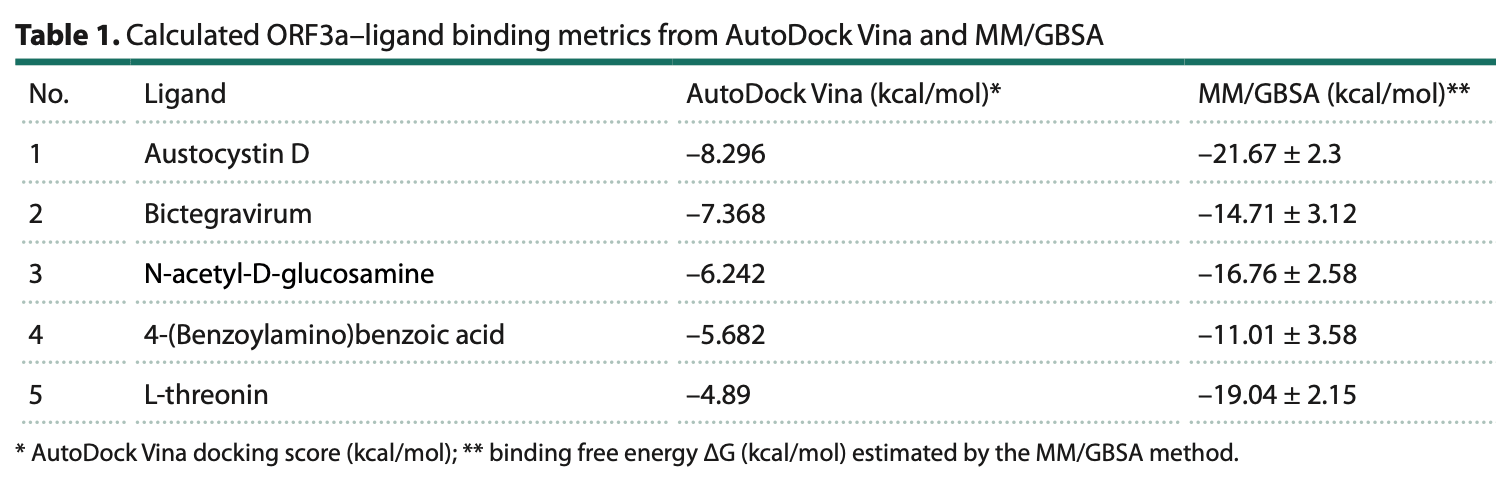
In silico study showing that five compounds may interact with the SARS-CoV-2 ORF3a protein using graph neural networks and molecular docking analysis. The binding sites of bictegravir and 4-(benzoylamino)benzoic acid were located on the cytosolic surface of ORF3a and partially overlapped with the ORF3a-VPS39 interaction region, suggesting these compounds could potentially disrupt the viral protein's ability to block autophagosome-lysosome fusion.
Ivanisenko et al., 12 Dec 2025, multiple countries, peer-reviewed, 4 authors.
Contact: itv@bionet.nsc.ru.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Prediction of interactions between the SARS-CoV-2 ORF3a protein and small-molecule ligands using the AND-System cognitive platform, graph neural networks, and molecular
Vavilov Journal of Genetics and Breeding, doi:10.18699/vjgb-25-113
In recent years, artificial intelligence methods based on the analysis of heterogeneous graphs of biomedical networks have become widely used for predicting molecular interactions. In particular, graph neural networks (GNNs) effectively identify missing edges in gene networks -such as protein-protein interaction, gene-disease, drug-target, and other networks -thereby enabling the prediction of new biological relationships. To reconstruct gene networks, cognitive systems for automatic text mining of scientific publications and databases are often employed. One such AI-driven platform, ANDSystem, is designed for automatic knowledge extraction of molecular interactions and, on this basis, the reconstruction of associative gene networks. The ANDSystem knowledge base contains information on more than 100 million interactions among diverse molecular genetic entities (genes, proteins, metabolites, drugs, etc.). The interactions span a wide range of types: regulatory relationships, physical interactions (protein-protein, protein-ligand), catalytic and chemical reactions, and associations among genes, phenotypes, diseases, and more. In the present study, we applied attention-based graph neural networks trained on the ANDSystem knowledge graph to predict new edges between proteins and ligands and to identify potential ligands for the SARS-CoV-2 ORF3a protein. The accessory protein ORF3a plays an important role in viral pathogenesis through ion-channel activity, induction of apoptosis, and the ability to modulate endolysosomal processes and the host innate immune response. Despite this broad functional spectrum, ORF3a has been explored far less as a pharmacological target than other viral proteins. Using a graph neural network, we predicted five small molecules of different origins (metabolites and a drug) that potentially interact with ORF3a: N-acetyl-D-glucosamine, 4-(benzoylamino)benzoic acid, austocystin D, bictegravirum, and L-threonine. Molecular docking and MM/GBSA affinity estimation indicate the potential ability of these compounds to form complexes with ORF3a. Localization analysis showed that the binding sites of bictegravir and 4-(benzoylamino)benzoic acid lie in a cytosolic surface pocket of the protein that is solvent-exposed; L-threonine binds within the intersubunit cleft of the dimer; and austocystin D and N-acetyl-D-glucosamine are positioned at the boundary between the cytosolic surface and the transmembrane region. The accessibility of these binding sites may be reduced by the influence of the lipid bilayer. The binding energetics for bictegravirum were more favorable than for 4-(benzoylamino)benzoic acid (docking score -7.37 kcal/mol; MM/GBSA ΔG -14.71 ± 3.12 kcal/mol), making bictegravirum a promising candidate for repurposing as an ORF3a inhibitor.
Conflict of interest. The authors declare no conflict of interest.
References
Ahsan, Sajib, Repurposing of approved drugs with potential to interact with SARS-CoV-2 receptor, Biochem Biophys Rep, doi:10.1016/j.bbrep.2021.100982
Ali, Alrashid, A review of methods for gene regulatory networks reconstruction and analysis, Artif Intell Rev, doi:10.1007/s10462-025-11257-z
Barberis, Timo, Amede, Vanella, Puricelli et al., Large-scale plasma analysis revealed new mechanisms and molecules associated with the host response to SARS-CoV-2, Int J Mol Sci, doi:10.3390/ijms21228623
Baysal, Ghafoor, Silme, Ignatov, Kniazeva, Molecular dynamics analysis of N-acetyl-D-glucosamine against specific SARSCoV2's pathogenicity factors, PLoS One, doi:10.1371/journal.pone.0252571
Boby, Fearon, Ferla, Filep, Koekemoer et al., Open science discovery of potent non-covalent SARS-CoV-2 main protease inhibitors, Science, doi:10.1126/science.abo7201
Bragina, Tiys, Freidin, Koneva, Demenkov et al., Insights into pathophysiology of dystropy through the analysis of gene networks: an example of bronchial asthma and tuberculosis, Immunogenetics, doi:10.1007/s00251-014-0786-1
Bragina, Tiys, Rudko, Ivanisenko, Freidin, Novel tuberculosis susceptibility candidate genes revealed by the reconstruction and analysis of associative networks, Infect Genet Evol, doi:10.1016/j.meegid.2016.10.030
Butikova, Basov, Rogachev, Gaisler, Ivanisenko et al., Metabolomic and gene networks approaches reveal the role of mitochondrial membrane proteins in response of human melanoma cells to THz radiation, Biochim Biophys Acta Mol Cell Biol Lipids, doi:10.1016/j.bbalip.2025.159595
Case, Aktulga, Belfon, Cerutti, Cisneros et al., None, J Chem Inf Model, doi:10.1021/acs.jcim.3c01153
Chicco, Jurman, The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation, BMC Genomics, doi:10.1186/s12864-019-6413-7
Dong, Galvan Achi, Du, Rong, Cui, Development of SARS-CoV-2 entry antivirals, Cell Insight, doi:10.1016/j.cellin.2023.100144
Eberhardt, Santos-Martins, Tillack, Forli, AutoDock Vina 1.2.0: new docking methods, expanded force field, and Python bindings, J Chem Inf Model, doi:10.1021/acs.jcim.1c00203
Fey, Lenssen, Fast graph representation learning with PyTorch Geometric, doi:10.48550/arXiv.1903.02428
Gallant, Thompson, Dejesus, Voskuhl, Wei et al., Antiviral activity, safety, and pharmacokinetics of bictegravir as 10-day monotherapy in HIV-1-infected adults, J Acquir Immune Defic Syndr, doi:10.1097/qai.0000000000001306
Gaudelet, Day, Jamasb, Soman, Regep et al., Utilizing graph machine learning within drug discovery and development, Brief Bioinform, doi:10.1093/bib/bbab159
Glorot, Bordes, Bengio, Deep sparse rectifier neural networks
Glotov, Tiys, Vashukova, Pakin, Demenkov et al., Molecular association of pathogenetic contributors to pre-eclampsia (pre-eclampsia associome), BMC Syst Biol, doi:10.1186/1752-0509-9-s2-s4
Gwon, Strand, Lindqvist, Nilsson, Saleeb et al., Antiviral activity of benzavir-2 against emerging flaviviruses, Viruses, doi:10.3390/v12030351
Harris, Primer on binary logistic regression, Fam Med Community Health, doi:10.1136/fmch-2021-001290
Hinkle, Trychta, Wires, Osborn, Leach et al., Subcellular localization of SARS-CoV-2 E and 3a proteins along the secretory pathway, J Mol Histol, doi:10.1007/s10735-025-10375-w
Islam, Strand, Saleeb, Svensson, Baranczewski et al., Anti-Rift Valley fever virus activity in vitro, pre-clinical pharmacokinetics and oral bioavailability of benzavir-2, a broad-acting antiviral compound, Sci Rep, doi:10.1038/s41598-018-20362-9
Ivanisenko, Demenkov, Ivanisenko, An accurate and efficient approach to knowledge extraction from scientific publications using structured ontology models, graph neural networks, and large language models, Int J Mol Sci, doi:10.3390/ijms252111811
Ivanisenko, Demenkov, Ivanisenko, Mishchenko, Saik, A new version of the ANDSystem tool for automatic extraction of knowledge from scientific publications with expanded functionality for reconstruction of associative gene networks by considering tissuespecific gene expression, BMC Bioinformatics, doi:10.1186/s12859-018-2567-6
Ivanisenko, Demenkov, Kolchanov, Ivanisenko, The new version of the ANDDigest tool with improved AI-based short names recognition, Int J Mol Sci, doi:10.3390/ijms232314934
Ivanisenko, Gaisler, Basov, Rogachev, Cheresiz et al., None, А. Иванисенко
Ivanisenko, Rogachev, Makarova, Basov, Gais Ler et al., AI assisted identification of primary and secondary metabolomic mar kers for postoperative delirium, Int J Mol Sci, doi:10.3390/ijms252111847
Ivanisenko, Saik, Demenkov, Ivanisenko, Savostianov et al., ANDDigest: a new webbased module of ANDSystem for the search of knowledge in the scientific literature, BMC Bioinformatics, doi:10.1186/s12859-020-03557-8
Ivanisenko, Saik, Ivanisenko, Tiys, Ivanisenko et al., ANDSystem: an associative network discovery system for automated literature mining in the field of biology, BMC Syst Biol, doi:10.1186/1752-0509-9-s2-s2
Kern, Sorum, Mali, Hoel, Sridharan et al., CryoEM structure of SARS-CoV-2 ORF3a in lipid nanodiscs, Nat Struct Mol Biol, doi:10.1038/s41594-021-00619-0
Krämer, Green, Pollard, Jr, Tugendreich, Causal analysis approaches in Ingenuity Pathway Analysis, Bioinformatics, doi:10.1093/bioinformatics/btt703
Larina, Pastushkova, Kh, Tiys, Kireev et al., Permanent proteins in the urine of healthy humans during the Mars-500 experiment, J Bioinform Comput Biol, doi:10.1142/s0219720015400016
Lebedeva, Gubarev, Mamardashvili, Zaitceva, Zdanovich et al., Theoretical and experimental study of interaction of macroheterocyclic compounds with ORF3a of SARS-CoV-2, Sci Rep, doi:10.1038/s41598-021-99072-8
Mao, Mohri, Zhong, Crossentropy loss functions: theoretical analysis and applications
Marks, Park, Arefolov, Russo, Ishihara et al., The selectivity of austocystin D arises from celllinespecific drug activation by cytochrome P450 enzymes, J Nat Prod, doi:10.1021/np100429s
Messina, Giombini, Montaldo, Sharma, Zoccoli et al., Looking for pathways related to COVID19: confirmation of pathogenic mechanisms by SARSCoV2host interactome, Cell Death Dis, doi:10.1038/s41419-021-03881-8
Metabase, Metacore, Early Research Intelligence Solutions
Miller, Houlihan, Matamala, Cabezas-Bratesco, Lee et al., The SARS-CoV-2 accessory protein Orf3a is not an ion channel, eLife, doi:10.7554/elife.84477
Momynaliev, Kashin, Chelysheva, Selezneva, Demina et al., Functional divergence of Helicobacter pylori related to early gastric cancer, J Proteome Res, doi:10.1021/pr900586w
Naqvi, Fatima, Muhammad, Fatima, Singh et al., Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies, Int J Biol Sci, doi:10.3389/fonc.2025.1575647
Ng, Correia, Seagal, Degoey, Schrimpf et al., Antiviral drug discovery for the treatment of COVID-19 infections, Viruses, doi:10.3390/v14050961
Nicholson, Greene, Constructing knowledge graphs and their biomedical applications, Comput Struct Biotechnol J, doi:10.1016/j.csbj.2020.05.017
Oner, Demirhan, Miraloglu, Yalin, Kurutas, Investigation of antiviral substances in COVID19 by molecular docking: in silico study, Afr Health Sci, doi:10.4314/ahs.v23i1.4
Páez-Franco, Torres-Ruiz, Sosa-Hernández, Cervantes-Díaz, Romero-Ramírez et al., Metabolomics analysis reveals a modified amino acid metabolism that correlates with altered oxygen homeostasis in COVID-19 patients, Sci Rep, doi:10.1038/s41598-021857880
Saik, Ivanisenko, Demenkov, Ivanisenko, Interactome of the hepatitis C virus: literature mining with ANDSystem, Virus Res, doi:10.1016/j.virusres.2015.12.003
Saik, Nimaev, Usmonov, Demenkov, Ivanisenko et al., Prioritization of genes involved in endothelial cell apoptosis by their implication in lymphedema using an analysis of associative gene networks with ANDSystem, BMC Med Genomics, doi:10.1186/s12920-019-0492-9
Sax, Arribas, Orkin, Lazzarin, Pozniak et al., GS US3801489 and GSUS3801490 study investigators. bictegravir/ emtricitabine/tenofovir alafenamide as initial treatment for HIV1: fiveyear follow-up from two randomized trials, EClinicalMedicine, doi:10.1016/j.eclinm.2023.101991
Stephens, Kunec, Henke, Vidal, Greishaber et al., The role of the tyrosine-based sorting signals of the ORF3a protein of SARS-CoV-2 in intracellular trafficking and pathogenesis, Viruses, doi:10.3390/v17040522
Stokes, Yang, Swanson, Cubillos-Ruiz, Donghia et al., A deep learning approach to antibiotic discovery, Cell, doi:10.1016/j.cell.2020.01.021
Sun, Zhang, Wei, Zheng, Zhao et al., Screening, simulation, and optimization design of small-molecule inhibitors of the SARS-CoV-2 spike glycoprotein, PLoS One, doi:10.1371/journal.pone.0245975
Szklarczyk, Kirsch, Koutrouli, Nastou, Mehryary et al., The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest, Nucleic Acids Res, doi:10.1093/nar/gkac1000
Tekin, Investigation of the effects of Nacetylglucosamine on the stability of the spike protein in SARS-CoV-2 by molecular dynamics simulations, Comput Theor Chem, doi:10.1016/j.comptc.2023.114049
Trott, Olson, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J Comput Chem, doi:10.1002/jcc.21334
Veličković, Cucurull, Casanova, Romero, Liò et al., Graph attention networks, doi:10.48550/arXiv.1710.10903
Voevoda, Kolchanov, Pokrovsky, Plasma metabolomics and gene regulatory networks analysis reveal the role of nonstructural SARS-CoV-2 viral proteins in metabolic dysregulation in COVID-19 patients, Sci Rep, doi:10.1038/s41598-022-24170-0
Von Delft, Hall, Kwong, Purcell, Saikatendu et al., Accelerating antiviral drug dis-Received September 25, Nat Rev Drug Discov, doi:10.1038/s41573-023-00692-8
Wu, Pan, Chen, Long, Zhang et al., A comprehensive survey on graph neural networks, IEEE Trans Neural Netw Learn Syst, doi:10.1109/TNNLS.2020.2978386
Zhang, Cruz-Cosme, Zhuang, Liu, Liu et al., A systemic and molecular study of subcellular localization of SARS-CoV-2 proteins, Signal Transduct Target Ther, doi:10.1038/s41392-021-00564-w
Zhang, Ejikemeuwa, Gerzanich, Nasr, Tang et al., Understanding the role of SARS-CoV-2 ORF3a in viral pathogenesis and COVID-19, Front Microbiol, doi:10.3389/fmicb.2022.854567
Zhang, Sun, Pei, Mao, Zhao et al., The SARS-CoV-2 protein ORF3a inhibits fusion of autophagosomes with lysosomes, Cell Discov, doi:10.1038/s41421-021-00268-z
Zhou, Xie, Lin, Yan, Towards understanding convergence and generalization of AdamW, IEEE Trans Pattern Anal Mach Intell, doi:10.1109/TPAMI.2024.3382294
Zitnik, Agrawal, Leskovec, Modeling polypharmacy side effects with graph convolutional networks, Bioinformatics, doi:10.1093/bioinformatics/bty294
DOI record:
{
"DOI": "10.18699/vjgb-25-113",
"ISSN": [
"2500-3259"
],
"URL": "http://dx.doi.org/10.18699/vjgb-25-113",
"abstract": "<jats:p> In recent years, artificial intelligence methods based on the analysis of heterogeneous graphs of biomedical networks have become widely used for predicting molecular interactions. In particular, graph neural networks (GNNs) effectively identify missing edges in gene networks – such as protein–protein interaction, gene–disease, drug–target, and other networks – thereby enabling the prediction of new biological relationships. To reconstruct gene networks, cognitive systems for automatic text mining of scientific publications and databases are often employed. One such AI-driven platform, ANDSystem, is designed for automatic knowledge extraction of molecular interactions and, on this basis, the reconstruction of associative gene networks. The ANDSystem knowledge base contains information on more than 100 million interactions among diverse molecular genetic entities (genes, proteins, metabolites, drugs, etc.). The interactions span a wide range of types: regulatory relationships, physical interactions (protein–protein, protein–ligand), catalytic and chemical reactions, and associations among genes, phenotypes, diseases, and more. In the present study, we applied attention-based graph neural networks trained on the ANDSystem knowledge graph to predict new edges between proteins and ligands and to identify potential ligands for the SARS-CoV-2 ORF3a protein. The accessory protein ORF3a plays an important role in viral pathogenesis through ion-channel activity, induction of apoptosis, and the ability to modulate endolysosomal processes and the host innate immune response. Despite this broad functional spectrum, ORF3a has been explored far less as a pharmacological target than other viral proteins. Using a graph neural network, we predicted five small molecules of different origins (metabolites and a drug) that potentially interact with ORF3a: N-acetyl-D-glucosamine, 4-(benzoylamino)benzoic acid, austocystin D, bictegravirum, and L-threonine. Molecular docking and MM/GBSA affinity estimation indicate the potential ability of these compounds to form complexes with ORF3a. Localization analysis showed that the binding sites of bictegravir and 4-(benzoylamino)benzoic acid lie in a cytosolic surface pocket of the protein that is solvent-exposed; L-threonine binds within the intersubunit cleft of the dimer; and austocystin D and N-acetyl-D-glucosamine are positioned at the boundary between the cytosolic surface and the transmembrane region. The accessibility of these binding sites may be reduced by the influence of the lipid bilayer. The binding energetics for bictegravirum were more favorable than for 4-(benzoylamino)benzoic acid (docking score −7.37 kcal/mol; MM/GBSA ΔG −14.71 ± 3.12 kcal/mol), making bictegravirum a promising candidate for repurposing as an ORF3a inhibitor.</jats:p>",
"author": [
{
"ORCID": "https://orcid.org/0000-0002-0005-9155",
"affiliation": [
{
"name": "Institute of Cytology and Genetics of the Siberian Branch of the Russian Academy of Sciences"
}
],
"authenticated-orcid": true,
"family": "Ivanisenko",
"given": "T. V.",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0000-0001-9433-8341",
"affiliation": [
{
"name": "Institute of Cytology and Genetics of the Siberian Branch of the Russian Academy of Sciences"
}
],
"authenticated-orcid": true,
"family": "Demenkov",
"given": "P. S.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-7537-2525",
"affiliation": [
{
"name": "Institute of Cytology and Genetics of the Siberian Branch of the Russian Academy of Sciences"
}
],
"authenticated-orcid": true,
"family": "Kleshchev",
"given": "M. A.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-1859-4631",
"affiliation": [
{
"name": "Institute of Cytology and Genetics of the Siberian Branch of the Russian Academy of Sciences"
}
],
"authenticated-orcid": true,
"family": "Ivanisenko",
"given": "V. A.",
"sequence": "additional"
}
],
"container-title": "Vavilov Journal of Genetics and Breeding",
"container-title-short": "Vestn. VOGiS",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
12,
12
]
],
"date-time": "2025-12-12T09:47:15Z",
"timestamp": 1765532835000
},
"deposited": {
"date-parts": [
[
2025,
12,
12
]
],
"date-time": "2025-12-12T09:48:22Z",
"timestamp": 1765532902000
},
"indexed": {
"date-parts": [
[
2025,
12,
12
]
],
"date-time": "2025-12-12T09:52:07Z",
"timestamp": 1765533127454,
"version": "3.48.0"
},
"is-referenced-by-count": 0,
"issue": "7",
"issued": {
"date-parts": [
[
2025,
12,
12
]
]
},
"journal-issue": {
"issue": "7",
"published-online": {
"date-parts": [
[
2025,
12,
12
]
]
}
},
"license": [
{
"URL": "https://vavilov.elpub.ru/jour/about/editorialPolicies#openAccessPolicy",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
12,
12
]
],
"date-time": "2025-12-12T00:00:00Z",
"timestamp": 1765497600000
}
}
],
"link": [
{
"URL": "https://vavilov.elpub.ru/jour/article/viewFile/4891/2021",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
}
],
"member": "7950",
"original-title": [],
"page": "1084-1096",
"prefix": "10.18699",
"published": {
"date-parts": [
[
2025,
12,
12
]
]
},
"published-online": {
"date-parts": [
[
2025,
12,
12
]
]
},
"publisher": "Institute of Cytology and Genetics, SB RAS",
"reference": [
{
"DOI": "10.1016/j.bbrep.2021.100982",
"doi-asserted-by": "crossref",
"key": "ref1",
"unstructured": "Ahsan T., Sajib A.A. Repurposing of approved drugs with potential to interact with SARS-CoV-2 receptor. Biochem Biophys Rep. 2021;26:100982. doi: 10.1016/j.bbrep.2021.100982"
},
{
"DOI": "10.1007/s10462-025-11257-z",
"doi-asserted-by": "crossref",
"key": "ref2",
"unstructured": "Ali S.I.M., Alrashid S.Z. A review of methods for gene regulatory networks reconstruction and analysis. Artif Intell Rev. 2025;58:256. doi: 10.1007/s1046202511257z"
},
{
"DOI": "10.3390/ijms21228623",
"doi-asserted-by": "crossref",
"key": "ref3",
"unstructured": "Barberis E., Timo S., Amede E., Vanella V.V., Puricelli C., Cappellano G., Raineri D., … Rolla R., Chiocchetti A., Baldanzi G., Marengo E., Manfredi M. Large-scale plasma analysis revealed new mechanisms and molecules associated with the host response to SARS-CoV-2. Int J Mol Sci. 2020;21(22):8623. doi: 10.3390/ijms21228623"
},
{
"DOI": "10.1371/journal.pone.0252571",
"doi-asserted-by": "crossref",
"key": "ref4",
"unstructured": "Baysal Ö., Abdul Ghafoor N., Silme R.S., Ignatov A.N., Kniazeva V. Molecular dynamics analysis of N-acetyl-D-glucosamine against specific SARSCoV2’s pathogenicity factors. PLoS One. 2021; 16(5):e0252571. doi: 10.1371/journal.pone.0252571"
},
{
"DOI": "10.1126/science.abo7201",
"doi-asserted-by": "crossref",
"key": "ref5",
"unstructured": "Boby M.L., Fearon D., Ferla M., Filep M., Koekemoer L., Robinson M.C., COVID Moonshot Consortium, … Zaidmann D., Zhang I., Zidane H., Zitzmann N., Zvornicanin S.N. Open science discovery of potent non-covalent SARS-CoV-2 main protease inhibitors. Science. 2023;380(6640):eabo7201. doi: 10.1126/science.abo7201"
},
{
"DOI": "10.1007/s00251-014-0786-1",
"doi-asserted-by": "crossref",
"key": "ref6",
"unstructured": "Bragina E.Y., Tiys E.S., Freidin M.B., Koneva L.A., Demenkov P.S., Ivanisenko V.A., Kolchanov N.A., Puzyrev V.P. Insights into pathophysiology of dystropy through the analysis of gene networks: an example of bronchial asthma and tuberculosis. Immunogenetics. 2014;66(78):457465. doi: 10.1007/s0025101407861"
},
{
"DOI": "10.1016/j.meegid.2016.10.030",
"doi-asserted-by": "crossref",
"key": "ref7",
"unstructured": "Bragina E.Y., Tiys E.S., Rudko A.A., Ivanisenko V.A., Freidin M.B. Novel tuberculosis susceptibility candidate genes revealed by the reconstruction and analysis of associative networks. Infect Genet Evol. 2016;46:118123. doi: 10.1016/j.meegid.2016.10.030"
},
{
"DOI": "10.1016/j.bbalip.2025.159595",
"doi-asserted-by": "crossref",
"key": "ref8",
"unstructured": "Butikova E.A., Basov N.V., Rogachev A.D., Gaisler E.V., Ivanisenko V.A., Demenkov P.S., Makarova A.L.A., … Pokrovsky A.G., Vinokurov N.A., Kanygin V.V., Popik V.M., Shevchenko O.A. Metabolomic and gene networks approaches reveal the role of mitochondrial membrane proteins in response of human melanoma cells to THz radiation. Biochim Biophys Acta Mol Cell Biol Lipids. 2025;1870(2):159595. doi: 10.1016/j.bbalip.2025.159595"
},
{
"DOI": "10.1021/acs.jcim.3c01153",
"doi-asserted-by": "crossref",
"key": "ref9",
"unstructured": "Case D.A., Aktulga H.M., Belfon K., Cerutti D.S., Andrés Cisneros G., Cruzeiro V.W.D., Forouzesh N., … Roitberg A., Simmerling C.S., York D.M., Nagan M.C., Merz K.M. Jr. AmberTools. J Chem Inf Model. 2023;63(20):61836191. doi: 10.1021/acs.jcim.3c01153"
},
{
"DOI": "10.1186/s12864-019-6413-7",
"doi-asserted-by": "crossref",
"key": "ref10",
"unstructured": "Chicco D., Jurman G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genomics. 2020;21:6. doi: 10.1186/s12864-019-64137"
},
{
"key": "ref11",
"unstructured": "Clarivate. MetaBase & MetaCore: Early Research Intelligence Solutions. Available at: http://clarivate.com/life-sciences-healthcare/research-development/discovery-development/early-research-intelligence-solutions/"
},
{
"DOI": "10.1016/j.cellin.2023.100144",
"doi-asserted-by": "crossref",
"key": "ref12",
"unstructured": "Dong M., Galvan Achi J.M., Du R., Rong L., Cui Q. Development of SARS-CoV-2 entry antivirals. Cell Insight. 2024;3(1):100144. doi: 10.1016/j.cellin.2023.100144"
},
{
"DOI": "10.1021/acs.jcim.1c00203",
"doi-asserted-by": "crossref",
"key": "ref13",
"unstructured": "Eberhardt J., Santos-Martins D., Tillack A.F., Forli S. AutoDock Vina 1.2.0: new docking methods, expanded force field, and Python bindings. J Chem Inf Model. 2021;61(8):38913898. doi: 10.1021/acs.jcim.1c00203"
},
{
"key": "ref14",
"unstructured": "Fey M., Lenssen J.E. Fast graph representation learning with PyTorch Geometric. arXiv. 2019. doi: 10.48550/arXiv.1903.02428"
},
{
"DOI": "10.1097/QAI.0000000000001306",
"doi-asserted-by": "crossref",
"key": "ref15",
"unstructured": "Gallant J.E., Thompson M., DeJesus E., Voskuhl G.W., Wei X., Zhang H., Martin H. Antiviral activity, safety, and pharmacokinetics of bictegravir as 10-day monotherapy in HIV-1-infected adults. J Acquir Immune Defic Syndr. 2017;75(1):6166. doi: 10.1097/QAI.0000000000001306"
},
{
"DOI": "10.1093/bib/bbab159",
"doi-asserted-by": "crossref",
"key": "ref16",
"unstructured": "Gaudelet T., Day B., Jamasb A.R., Soman J., Regep C., Liu G., Hayter J.B.R., Vickers R., Roberts C., Tang J., Roblin D., Blundell T.L., Bronstein M.M., Taylor-King J.P. Utilizing graph machine learning within drug discovery and development. Brief Bioinform. 2021; 22(6):bbab159. doi: 10.1093/bib/bbab159"
},
{
"key": "ref17",
"unstructured": "Glorot X., Bordes A., Bengio Y. Deep sparse rectifier neural networks. In: Proceedings of the Fourteenth International Conference on Artificial Intelligence and Statistics (AISTATS). 2011;315323. Available at: https://proceedings.mlr.press/v15/glorot11a/glorot11a.pdf"
},
{
"DOI": "10.1186/1752-0509-9-S2-S4",
"doi-asserted-by": "crossref",
"key": "ref18",
"unstructured": "Glotov A.S., Tiys E.S., Vashukova E.S., Pakin V.S., Demenkov P.S., Saik O.V., Ivanisenko T.V., Arzhanova O.N., Mozgovaya E.V., Zainulina M.S., Kolchanov N.A., Baranov V.S., Ivanisenko V.A. Molecular association of pathogenetic contributors to pre-eclampsia (pre-eclampsia associome). BMC Syst Biol. 2015;9(Suppl. 2):S4. doi: 10.1186/175205099S2S4"
},
{
"DOI": "10.3390/v12030351",
"doi-asserted-by": "crossref",
"key": "ref19",
"unstructured": "Gwon Y.D., Strand M., Lindqvist R., Nilsson E., Saleeb M., Elofsson M., Överby A.K., Evander M. Antiviral activity of benzavir-2 against emerging flaviviruses. Viruses. 2020;12(3):351. doi: 10.3390/v12030351"
},
{
"DOI": "10.1136/fmch-2021-001290",
"doi-asserted-by": "crossref",
"key": "ref20",
"unstructured": "Harris J.K. Primer on binary logistic regression. Fam Med Community Health. 2021;9(Suppl. 1):e001290. doi 10.1136/fmch-2021-001290"
},
{
"DOI": "10.1007/s10735-025-10375-w",
"doi-asserted-by": "crossref",
"key": "ref21",
"unstructured": "Hinkle J.J., Trychta K.A., Wires E.S., Osborn R.M., Leach J.R., Faraz Z.F., Svarcbahs R., Richie C.T., Dewhurst S., Harvey B.K. Subcellular localization of SARS-CoV-2 E and 3a proteins along the secretory pathway. J Mol Histol. 2025;56(2):98. doi: 10.1007/s1073502510375w"
},
{
"DOI": "10.1038/s41598-018-20362-9",
"doi-asserted-by": "crossref",
"key": "ref22",
"unstructured": "Islam M., Strand M., Saleeb M., Svensson R., Baranczewski P., Artursson P., Wadell G., Ahlm C., Elofsson M., Evander M. Anti-Rift Valley fever virus activity in vitro, pre-clinical pharmacokinetics and oral bioavailability of benzavir-2, a broad-acting antiviral compound. Sci Rep. 2018;8:1925. doi: 10.1038/s41598-018-20362-9"
},
{
"DOI": "10.1186/s12859-020-03557-8",
"doi-asserted-by": "crossref",
"key": "ref23",
"unstructured": "Ivanisenko T.V., Saik O.V., Demenkov P.S., Ivanisenko N.V., Savostianov A.N., Ivanisenko V.A. AND-Digest: a new webbased module of AND-System for the search of knowledge in the scientific literature. BMC Bioinformatics. 2020;21(Suppl 11):228. doi: 10.1186/s12859-020035578"
},
{
"DOI": "10.3390/ijms232314934",
"doi-asserted-by": "crossref",
"key": "ref24",
"unstructured": "Ivanisenko T.V., Demenkov P.S., Kolchanov N.A., Ivanisenko V.A. The new version of the AND-Digest tool with improved AI-based short names recognition. Int J Mol Sci. 2022;23(23):14934. doi: 10.3390/ijms232314934"
},
{
"DOI": "10.3390/ijms252111811",
"doi-asserted-by": "crossref",
"key": "ref25",
"unstructured": "Ivanisenko T.V., Demenkov P.S., Ivanisenko V.A. An accurate and efficient approach to knowledge extraction from scientific publications using structured ontology models, graph neural networks, and large language models. Int J Mol Sci. 2024;25(21):11811. doi: 10.3390/ijms252111811"
},
{
"DOI": "10.1186/1752-0509-9-S2-S2",
"doi-asserted-by": "crossref",
"key": "ref26",
"unstructured": "Ivanisenko V.A., Saik O.V., Ivanisenko N.V., Tiys E.S., Ivanisenko T.V., Demenkov P.S., Kolchanov N.A. ANDSystem: an associative network discovery system for automated literature mining in the field of biology. BMC Syst Biol. 2015;9(Suppl. 2):S2. doi: 10.1186/17520509-9-S2-S2"
},
{
"DOI": "10.1186/s12859-018-2567-6",
"doi-asserted-by": "crossref",
"key": "ref27",
"unstructured": "Ivanisenko V.A., Demenkov P.S., Ivanisenko T.V., Mishchenko E.L., Saik O.V. A new version of the AND-System tool for automatic extraction of knowledge from scientific publications with expanded functionality for reconstruction of associative gene networks by considering tissuespecific gene expression. BMC Bioinformatics. 2019; 20(1):34. doi: 10.1186/s1285901825676"
},
{
"DOI": "10.1038/s41598-022-24170-0",
"doi-asserted-by": "crossref",
"key": "ref28",
"unstructured": "Ivanisenko V.A., Gaisler E.V., Basov N.V., Rogachev A.D., Cheresiz S.V., Ivanisenko T.V., Demenkov P.S., … Karpenko T.N., Velichko A.J., Voevoda M.I., Kolchanov N.A., Pokrovsky A.G. Plasma metabolomics and gene regulatory networks analysis reveal the role of nonstructural SARS-CoV-2 viral proteins in metabolic dysregulation in COVID-19 patients. Sci Rep. 2022;12(1):19977. doi: 10.1038/s41598022241700"
},
{
"DOI": "10.3390/ijms252111847",
"doi-asserted-by": "crossref",
"key": "ref29",
"unstructured": "Ivanisenko V.A., Rogachev A.D., Makarova A.A., Basov N.V., Gaisler E.V., Kuzmicheva I.N., Demenkov P.S., … Kolchanov N.A., Plesko V.V., Moroz G.B., Lomivorotov V.V., Pokrovsky A.G. AIassisted identification of primary and secondary metabolomic markers for postoperative delirium. Int J Mol Sci. 2024;25(21):11847. doi: 10.3390/ijms252111847"
},
{
"DOI": "10.1038/s41594-021-00619-0",
"doi-asserted-by": "crossref",
"key": "ref30",
"unstructured": "Kern D.M., Sorum B., Mali S.S., Hoel C.M., Sridharan S., Remis J.P., Toso D.B., Kotecha A., Bautista D.M., Brohawn S.G. CryoEM structure of SARS-CoV-2 ORF3a in lipid nanodiscs. Nat Struct Mol Biol. 2021;28(7):573582. doi: 10.1038/s41594-021-00619-0"
},
{
"DOI": "10.1093/bioinformatics/btt703",
"doi-asserted-by": "crossref",
"key": "ref31",
"unstructured": "Krämer A., Green J., Pollard J. Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014; 30(4):523530. doi: 10.1093/bioinformatics/btt703"
},
{
"DOI": "10.1142/S0219720015400016",
"doi-asserted-by": "crossref",
"key": "ref32",
"unstructured": "Larina I.M., Pastushkova L.Kh., Tiys E.S., Kireev K.S., Kononikhin A.S., Starodubtseva N.L., Popov I.A., Custaud M.-A., Dobrokhotov I.V., Nikolaev E.N., Kolchanov N.A., Ivanisenko V.A. Permanent proteins in the urine of healthy humans during the Mars-500 experiment. J Bioinform Comput Biol. 2015;13(1):1540001. doi: 10.1142/S0219720015400016"
},
{
"DOI": "10.1038/s41598-021-99072-8",
"doi-asserted-by": "crossref",
"key": "ref33",
"unstructured": "Lebedeva N.S., Gubarev Y.A., Mamardashvili G.M., Zaitceva S.V., Zdanovich S.A., Malyasova A.S., Romanenko J.V., Koifman M.O., Koifman O.I. Theoretical and experimental study of interaction of macroheterocyclic compounds with ORF3a of SARS-CoV-2. Sci Rep. 2021;11:19481. doi: 10.1038/s41598021990728"
},
{
"key": "ref34",
"unstructured": "Mao A., Mohri M., Zhong Y. Crossentropy loss functions: theoretical analysis and applications. In: International Conference on Machine Learning (ICML). 2023;23803-23828. Available at: https://proceedings.mlr.press/v202/mao23b/mao23b.pdf"
},
{
"DOI": "10.1021/np100429s",
"doi-asserted-by": "crossref",
"key": "ref35",
"unstructured": "Marks K.M., Park E.S., Arefolov A., Russo K., Ishihara K., Ring J.E., Clardy J., Clarke A.S., Pelish E.P. The selectivity of austocystin D arises from celllinespecific drug activation by cytochrome P450 enzymes. J Nat Prod. 2011;74(4):567573. doi: 10.1021/np100429s"
},
{
"DOI": "10.1038/s41419-021-03881-8",
"doi-asserted-by": "crossref",
"key": "ref36",
"unstructured": "Messina F., Giombini E., Montaldo C., Sharma A.A., Zoccoli A., Sekaly R.P., Locatelli F., Zumla A., Maeurer M., Capobianchi M.R., Lauria F.N., Ippolito G. Looking for pathways related to COVID19: confirmation of pathogenic mechanisms by SARSCoV2host interactome. Cell Death Dis. 2021;12(8):788. doi: 10.1038/s41419-021-03881-8"
},
{
"DOI": "10.7554/eLife.84477",
"doi-asserted-by": "crossref",
"key": "ref37",
"unstructured": "Miller A.N., Houlihan P.R., Matamala E., Cabezas-Bratesco D., Lee G.Y., CristoforiArmstrong B., Dilan T.L., SanchezMartinez S., Matthies D., Yan R., Yu Z., Ren D., Brauchi S.E., Clapham D.E. The SARS-CoV-2 accessory protein Orf3a is not an ion channel. eLife. 2023;12:e84477. doi: 10.7554/eLife.84477"
},
{
"DOI": "10.1021/pr900586w",
"doi-asserted-by": "crossref",
"key": "ref38",
"unstructured": "Momynaliev K.T., Kashin S.V., Chelysheva V.V., Selezneva O.V., Demina I.A., Serebryakova M.V., Alexeev D., Ivanisenko V.A., Aman E., Govorun V.M. Functional divergence of Helicobacter pylori related to early gastric cancer. J Proteome Res. 2010;9(1): 254267. doi: 10.1021/pr900586w"
},
{
"DOI": "10.1016/j.bbadis.2020.165878",
"doi-asserted-by": "crossref",
"key": "ref39",
"unstructured": "Naqvi A.A.T., Fatima K., Muhammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies. Int J Biol Sci. 2020;16(10):17081724. doi: 10.7150/ijbs.45127"
},
{
"DOI": "10.3390/v14050961",
"doi-asserted-by": "crossref",
"key": "ref40",
"unstructured": "Ng T.I., Correia I., Seagal J., DeGoey D.A., Schrimpf M.R., Hardee D.J., Noey E.L., Kati W.M. Antiviral drug discovery for the treatment of COVID-19 infections. Viruses. 2022;14(5):961. doi: 10.3390/v14050961"
},
{
"DOI": "10.1016/j.csbj.2020.05.017",
"doi-asserted-by": "crossref",
"key": "ref41",
"unstructured": "Nicholson D.N., Greene C.S. Constructing knowledge graphs and their biomedical applications. Comput Struct Biotechnol J. 2020;18: 1414-1421. doi: 10.1016/j.csbj.2020.05.017"
},
{
"DOI": "10.4314/ahs.v23i1.4",
"doi-asserted-by": "crossref",
"key": "ref42",
"unstructured": "Oner E., Demirhan I., Miraloglu M., Yalin S., Kurutas E.B. Investigation of antiviral substances in COVID19 by molecular docking: in silico study. Afr Health Sci. 2023;23(1):2336. doi: 10.4314/ahs.v23i1.4"
},
{
"DOI": "10.1038/s41598-021-85788-0",
"doi-asserted-by": "crossref",
"key": "ref43",
"unstructured": "Páez-Franco J.C., Torres-Ruiz J., Sosa-Hernández V.A., Cervantes-Díaz R., Romero-Ramírez S., Pérez-Fragoso A., Meza-Sánchez D.E., GermánAcacio J.M., MaravillasMontero J.L., MejíaDomínguez N.R., PoncedeLeón A., UlloaAguirre A., GómezMartín D., Llorente L. Metabolomics analysis reveals a modified amino acid metabolism that correlates with altered oxygen homeostasis in COVID-19 patients. Sci Rep. 2021;11(1):6350. doi: 10.1038/s41598-021857880"
},
{
"DOI": "10.1016/j.virusres.2015.12.003",
"doi-asserted-by": "crossref",
"key": "ref44",
"unstructured": "Saik O.V., Ivanisenko T.V., Demenkov P.S., Ivanisenko V.A. Interactome of the hepatitis C virus: literature mining with AND-System. Virus Res. 2016;218:4048. doi: 10.1016/j.virusres.2015.12.003"
},
{
"DOI": "10.1186/s12920-019-0492-9",
"doi-asserted-by": "crossref",
"key": "ref45",
"unstructured": "Saik O.V., Nimaev V.V., Usmonov D.B., Demenkov P.S., Ivanisenko T.V., Lavrik I.N., Ivanisenko V.A. Prioritization of genes involved in endothelial cell apoptosis by their implication in lymphedema using an analysis of associative gene networks with AND-System. BMC Med Genomics. 2019;12(Suppl. 2):117. doi: 10.1186/s12920-019-0492-9"
},
{
"DOI": "10.1016/j.eclinm.2023.101991",
"doi-asserted-by": "crossref",
"key": "ref46",
"unstructured": "Sax P.E., Arribas J.R., Orkin C., Lazzarin A., Pozniak A., DeJesus E., Maggiolo F., … Hindman J.T., Martin H., Baeten J.M., Wohl D.; GS US3801489 and GSUS3801490 study investigators. bictegravir/emtricitabine/tenofovir alafenamide as initial treatment for HIV1: fiveyear follow-up from two randomized trials. EClinicalMedicine. 2023;59:101991. doi: 10.1016/j.eclinm.2023.101991"
},
{
"DOI": "10.3390/v17040522",
"doi-asserted-by": "crossref",
"key": "ref47",
"unstructured": "Stephens E.B., Kunec D., Henke W., Vidal R.M., Greishaber B., Saud R., Kalamvoki M., Singh G., Kafle S., Trujillo J.D., Ferreyra F.M., Morozov I., Richt J.A. The role of the tyrosine-based sorting signals of the ORF3a protein of SARS-CoV-2 in intracellular trafficking and pathogenesis. Viruses. 2025;17(4):522. doi: 10.3390/v17040522"
},
{
"DOI": "10.1016/j.cell.2020.01.021",
"doi-asserted-by": "crossref",
"key": "ref48",
"unstructured": "Stokes J.M., Yang K., Swanson K., Jin W., Cubillos-Ruiz A., Donghia N.M., MacNair C.R., … Church G.M., Brown E.D., Jaakkola T.S., Barzilay R., Collins J.J. A deep learning approach to antibiotic discovery. Cell. 2020;180(4):688702. doi: 10.1016/j.cell.2020.01.021"
},
{
"DOI": "10.1371/journal.pone.0245975",
"doi-asserted-by": "crossref",
"key": "ref49",
"unstructured": "Sun C., Zhang J., Wei J., Zheng X., Zhao X., Fang Z., Xu D., Yuan H., Liu Y. Screening, simulation, and optimization design of small-molecule inhibitors of the SARS-CoV-2 spike glycoprotein. PLoS One. 2021;16(1):e0245975. doi: 10.1371/journal.pone.0245975"
},
{
"DOI": "10.1093/nar/gkac1000",
"doi-asserted-by": "crossref",
"key": "ref50",
"unstructured": "Szklarczyk D., Kirsch R., Koutrouli M., Nastou K., Mehryary F., Hachilif R., Gable A.L., Fang T., Doncheva N.T., Pyysalo S., Bork P., Jensen L.J., von Mering C. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023; 51(D1):D638D646. doi: 10.1093/nar/gkac1000"
},
{
"DOI": "10.1016/j.comptc.2023.114049",
"doi-asserted-by": "crossref",
"key": "ref51",
"unstructured": "Tekin E.D. Investigation of the effects of Nacetylglucosamine on the stability of the spike protein in SARS-CoV-2 by molecular dynamics simulations. Comput Theor Chem. 2023;1222:114049. doi: 10.1016/j.comptc.2023.114049"
},
{
"DOI": "10.1002/jcc.21334",
"doi-asserted-by": "crossref",
"key": "ref52",
"unstructured": "Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455461. doi: 10.1002/jcc.21334"
},
{
"DOI": "10.1002/cpu.30542",
"doi-asserted-by": "crossref",
"key": "ref53",
"unstructured": "U.S. Food and Drug Administration (FDA). FDA approves first treatment for COVID-19 (Veklury/remdesivir): press release. 22 Oct 2020. Available at: https://www.fda.gov/news-events/press-announcements/fda-approves-first-treatment-covid-19"
},
{
"key": "ref54",
"unstructured": "U.S. Food and Drug Administration. FDA approves first oral antiviral for treatment of COVID-19 in adults: press announcement. 20230525. Available at: https://www.fda.gov/news-events/press-announcements/fda-approves-first-oral-antiviral-treatment-covid-19-adults"
},
{
"key": "ref55",
"unstructured": "Veličković P., Cucurull G., Casanova A., Romero A., Liò P., Bengio Y. Graph attention networks. arXiv. 2017. doi: 10.48550/arXiv.1710.10903"
},
{
"DOI": "10.1038/s41573-023-00692-8",
"doi-asserted-by": "crossref",
"key": "ref56",
"unstructured": "von Delft A., Hall M.D., Kwong A.D., Purcell L.A., Saikatendu K.S., Schmitz U., Tallarico J.A., Lee A.A. Accelerating antiviral drug discovery: lessons from COVID-19. Nat Rev Drug Discov. 2023;22(7): 585-603. doi: 10.1038/s41573023006928"
},
{
"DOI": "10.1109/TNNLS.2020.2978386",
"doi-asserted-by": "crossref",
"key": "ref57",
"unstructured": "Wu Z., Pan S., Chen F., Long G., Zhang C., Yu P.S. A comprehensive survey on graph neural networks. IEEE Trans Neural Netw Learn Syst. 2021;32(1):424. doi: 10.1109/TNNLS.2020.2978386"
},
{
"DOI": "10.1038/s41392-021-00564-w",
"doi-asserted-by": "crossref",
"key": "ref58",
"unstructured": "Zhang J., Cruz-Cosme R., Zhuang M.W., Liu D., Liu Y., Teng S., Wang P.H., Tang Q. A systemic and molecular study of subcellular localization of SARS-CoV-2 proteins. Signal Transduct Target Ther. 2021;6(1):192. doi: 10.1038/s41392-021-00564-w"
},
{
"DOI": "10.3389/fmicb.2022.854567",
"doi-asserted-by": "crossref",
"key": "ref59",
"unstructured": "Zhang J., Ejikemeuwa A., Gerzanich V., Nasr M., Tang Q., Simard J.M., Zhao R.Y. Understanding the role of SARS-CoV-2 ORF3a in viral pathogenesis and COVID-19. Front Microbiol. 2022;13:854567. doi: 10.3389/fmicb.2022.854567"
},
{
"DOI": "10.1038/s41421-021-00268-z",
"doi-asserted-by": "crossref",
"key": "ref60",
"unstructured": "Zhang Y., Sun H., Pei R., Mao B., Zhao Z., Li H., Lin Y., Lu K. The SARS-CoV-2 protein ORF3a inhibits fusion of autophagosomes with lysosomes. Cell Discov. 2021;7:31. doi: 10.1038/s41421-021-00268-z"
},
{
"DOI": "10.1109/TPAMI.2024.3382294",
"doi-asserted-by": "crossref",
"key": "ref61",
"unstructured": "Zhou P., Xie X., Lin Z., Yan S. Towards understanding convergence and generalization of AdamW. IEEE Trans Pattern Anal Mach Intell. 2024;46(9):64866493. doi: 10.1109/TPAMI.2024.3382294"
},
{
"DOI": "10.1093/bioinformatics/bty294",
"doi-asserted-by": "crossref",
"key": "ref62",
"unstructured": "Zitnik M., Agrawal M., Leskovec J. Modeling polypharmacy side effects with graph convolutional networks. Bioinformatics. 2018; 34(13):i457i466. doi: 10.1093/bioinformatics/bty294"
}
],
"reference-count": 62,
"references-count": 62,
"relation": {},
"resource": {
"primary": {
"URL": "https://vavilov.elpub.ru/jour/article/view/4891"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Prediction of interactions between the SARS-CoV-2 ORF3a protein and small-molecule ligands using the AND-System cognitive platform, graph neural networks, and molecular",
"type": "journal-article",
"volume": "29"
}