Host Immune Response Profiling for the Diagnosis of Infectious Diseases
et al., The Journal of Infectious Diseases, doi:10.1093/infdis/jiaf428, Aug 2025
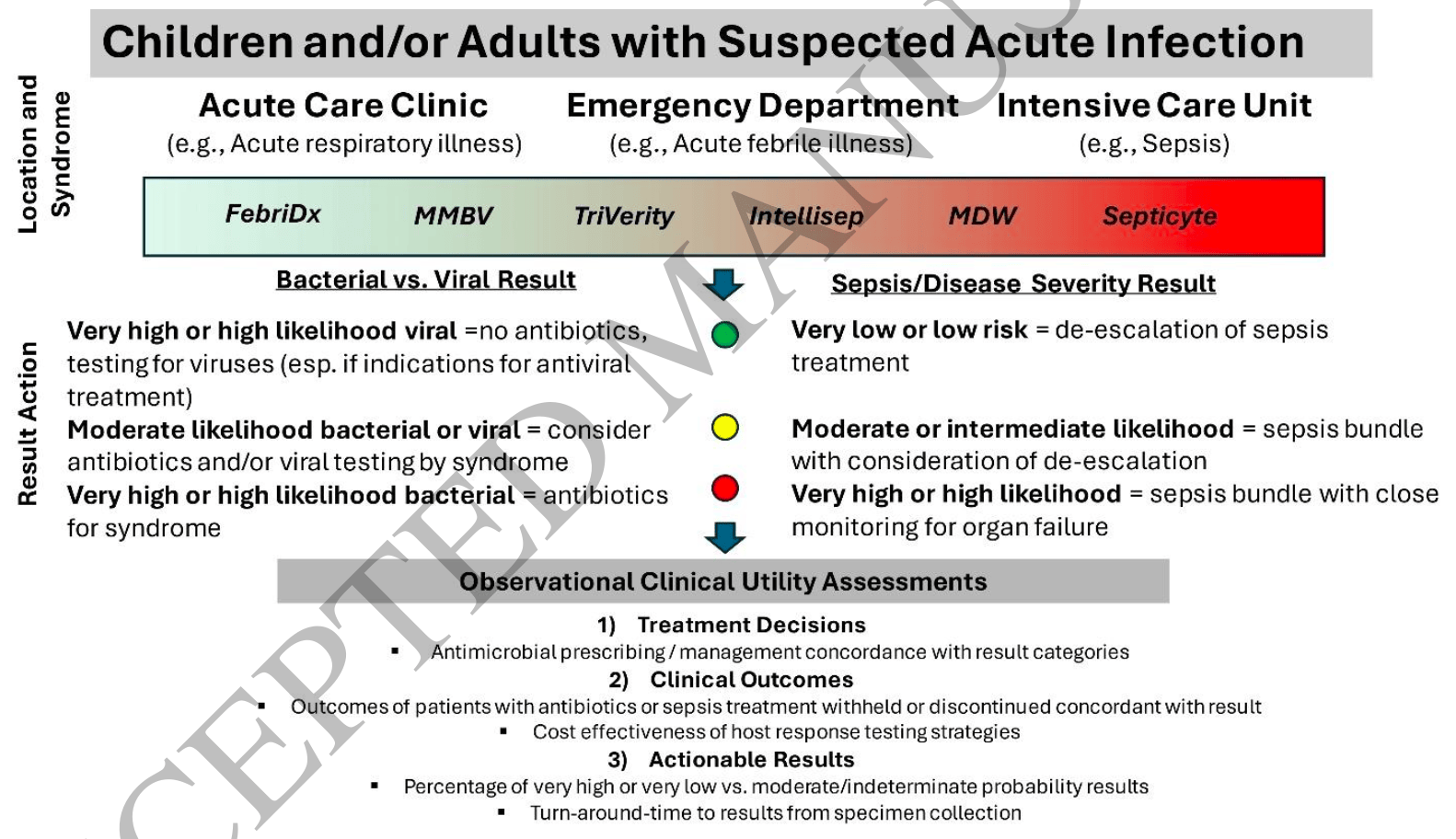
Review of host immune response (HR) diagnostics for infectious diseases, which measure gene expression profiles or inflammatory protein concentrations to help distinguish between bacterial and viral infections. Authors note that FebriDx has shown good performance for COVID-19 (with 91% sensitivity, 92% specificity, and 97% negative predictive value for SARS-CoV-2). HR diagnostics may be particularly valuable when dealing with novel pathogens or when other testing is unavailable.
Hanson et al., 11 Aug 2025, retrospective, USA, peer-reviewed, 2 authors.
Contact: kim.hanson@hsc.utah.edu, e.t@duke.edu.
Host Immune Response Profiling for the Diagnosis of Infectious Diseases
The Journal of Infectious Diseases, doi:10.1093/infdis/jiaf428
Recent advances in infectious diseases diagnostics include the development, validation, and commercialization of new tests that measure host gene expression profiles or inflammatory protein concentrations. Interrogating host immune responses may help separate infectious from noninfectious inflammation, differentiate infection types and/or predict sepsis severity/subtype. This review summarizes the current state-of-the art in host response (HR) diagnostics for infectious diseases with a focus on test accuracy. Few studies have assessed the potential impact of HR testing on patient outcomes. We summarize current clinical evidence gaps and describe the types of studies needed to inform optimal integration in clinical practice.
References
Agnello, Evaluating monocyte distribution width in pediatric emergency care, Clin Chim Acta
Antcliffe, Transcriptomic Signatures in Sepsis and a Differential Response to Steroids. From the VANISH Randomized Trial, Am J Respir Crit Care Med
Bachur, A rapid host-protein test for differentiating bacterial from viral infection: Apollo diagnostic accuracy study, J Am Coll Emerg Physicians Open
Bakochi, Cerebrospinal fluid proteome maps detect pathogen-specific host response patterns in meningitis, Elife
Balk, Rapid and Robust Identification of Sepsis Using SeptiCyte RAPID in a Heterogeneous Patient Population, J Clin Med
Balk, Validation of SeptiCyte RAPID to Discriminate Sepsis from Non-Infectious Systemic Inflammation, J Clin Med
Benoit, Seven-year performance of a clinical metagenomic next-generation sequencing test for diagnosis of central nervous system infections, Nat Med
Crawford, Rapid Biophysical Analysis of Host Immune Cell Variations Associated with Sepsis, Am J Respir Crit Care Med
Damschroder, The updated Consolidated Framework for Implementation Research based on user feedback, Implementation Science
Davenport, Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study, Lancet Respir Med
Dedeoglu, Comparison of two rapid host-response tests for distinguishing bacterial and viral infection in adults with acute respiratory infection, J Infect
Di Carlo, A mechanical biomarker of cell state in medicine, J Lab Autom
Dunning, Progression of whole-blood transcriptional signatures from interferon-induced to neutrophil-associated patterns in severe influenza, Nat Immunol
Fda, None, MeMed BV
Fda, None, SeptiCyte RAPID
Fda, Unicel DxH 800 Cellular Analysis System with Early Sepsis Indicator Application 510(k) Number K
García-Álvarez, Monocyte distribution width (MDW): study of reference values in blood donors, Clin Chem Lab Med
Glaser, In search of encephalitis etiologies: diagnostic challenges in the California Encephalitis Project, 1998-2000, Clin Infect Dis
Goto, Infectious Disease-Related Emergency Department Visits of Elderly Adults in the United States, 2011-2012, J Am Geriatr Soc
Haller, Kochs, Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity, J Interferon Cytokine Res
Hasegawa, Infectious Disease-related Emergency Department Visits Among Children in the US, Pediatr Infect Dis J
He, The Optimization and Biological Significance of a 29-Host-Immune-mRNA Panel for the Diagnosis of Acute Infections and Sepsis, J Pers Med
Holzinger, Induction of MxA gene expression by influenza A virus requires type I or type III interferon signaling, J Virol
Hou, Monocyte Distribution Width in Children With Systemic Inflammatory Response: Retrospective Cohort Examining Association With Early Sepsis, Pediatr Crit Care Med
Iglesias-Ussel, A Rapid Host Response Blood Test for Bacterial/Viral Infection Discrimination Using a Portable Molecular Diagnostic Platform, Open Forum Infect Dis
Jackson, A multi-platform approach to identify a blood-based host protein signature for distinguishing between bacterial and viral infections in febrile children (PERFORM): a multicohort machine learning study, The Lancet Digital Health
Kalantar, Integrated host-microbe plasma metagenomics for sepsis diagnosis in a prospective cohort of critically ill adults, Nat Microbiol
Kalmovich, Implementation of a rapid host-protein diagnostic test for distinguishing bacterial and viral infections in adults presenting to urgent care centers: a pragmatic cohort study, BMC Med
Kalmovich, Rahamim-Cohen, Shapiro Ben, Impact on Patient Management of a Novel Host Response Test for Distinguishing Bacterial and Viral Infections: Real World Evidence from the Urgent Care Setting, Biomedicines
Khera, Race and Gender Differences in C-Reactive Protein Levels, JACC
King, Fleming-Dutra, Hicks, Advances in optimizing the prescription of antibiotics in outpatient settings, Bmj
Klein, Diagnostic Accuracy of a Real-Time Host-Protein Test for Infection, Pediatrics
Ko, Prospective Validation of a Rapid Host Gene Expression Test to Discriminate Bacterial From Viral Respiratory Infection, JAMA Netw Open
Langelier, Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults, Proc Natl Acad Sci U S A
Lippi, FebriDx for rapid screening of patients with suspected COVID-19 upon hospital admission: systematic literature review and meta-analysis, J Hosp Infect
Malinovska, Monocyte Distribution Width as a Diagnostic Marker for Infection: A Systematic Review and Meta-analysis, Chest
Malinovska, Monocyte distribution width as part of a broad pragmatic sepsis screen in the emergency department, J Am Coll Emerg Physicians Open
Maslove, Redefining critical illness, Nat Med
Maslove, Wong, Gene expression profiling in sepsis: timing, tissue, and translational considerations, Trends Mol Med
Mayhew, A generalizable 29-mRNA neural-network classifier for acute bacterial and viral infections, Nat Commun
Mick, Integrated host/microbe metagenomics enables accurate lower respiratory tract infection diagnosis in critically ill children, J Clin Invest
Mogensen, Pathogen recognition and inflammatory signaling in innate immune defenses, Clin Microbiol Rev
Nijman, A Novel Framework for Phenotyping Children With Suspected or Confirmed Infection for Future Biomarker Studies, Front Pediatr
Novak, MeMed BV testing in emergency department patients presenting with febrile illness concerning for respiratory tract infection, Am J Emerg Med
O'neal, Jr, Cellular host response sepsis test for risk stratification of patients in the emergency department: A pooled analysis, Acad Emerg Med
Oliver Liesenfeld, Thomas Aufderheide, Clements, Devos, Fischer et al., Rapid and Accurate Diagnosis and Prognosis of Acute Infections and Sepsis from Whole Blood Using Host Response mRNA amplification and Result Interpretation by Machine -Learning Classifiers
Omura, Host Response Profiling from Clinical Metagenomic Sequencing Data for Diagnosis of Central Nervous System Infections, Open Forum Infectious Diseases
Oved, A novel host-proteome signature for distinguishing between acute bacterial and viral infections, PLoS One
Papan, Combinatorial Host-Response Biomarker Signature (BV Score) and Its Subanalytes TRAIL, IP-10, and C-Reactive Protein in Children With Mycoplasma pneumoniae Community-Acquired Pneumonia, J Infect Dis
Patel, Clinically Adjudicated Reference Standards for Evaluation of Infectious Diseases Diagnostics, Clin Infect Dis
Proctor, Outcomes for implementation research: conceptual distinctions, measurement challenges, and research agenda, Adm Policy Ment Health
Ramachandran, Integrating central nervous system metagenomics and host response for diagnosis of tuberculosis meningitis and its mimics, Nat Commun
Rao, A robust host-response-based signature distinguishes bacterial and viral infections across diverse global populations, Cell Rep Med
Shapiro, Diagnostic Accuracy of a Bacterial and Viral Biomarker Point-of-Care Test in the Outpatient Setting, JAMA Netw Open
Sweeney, A comprehensive time-course-based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set, Sci Transl Med
Sweeney, Wong, Khatri, Robust classification of bacterial and viral infections via integrated host gene expression diagnostics, Sci Transl Med
Van Amstel, Clinical subtypes in critically ill patients with sepsis: validation and parsimonious classifier model development, Crit Care
Van Amstel, Uncovering heterogeneity in sepsis: a comparative analysis of subphenotypes, Intensive Care Med
Van Houten, Expert panel diagnosis demonstrated high reproducibility as reference standard in infectious diseases, J Clin Epidemiol
Wilcox, Use of the FebriDx® host-response point-of-care test may reduce antibiotic use for respiratory tract infections in primary care: a mixed -methods feasibility study, J Antimicrob Chemother
Woods, A host transcriptional signature for presymptomatic detection of infection in humans exposed to influenza H1N1 or H3N2, PLoS One
DOI record:
{
"DOI": "10.1093/infdis/jiaf428",
"ISSN": [
"0022-1899",
"1537-6613"
],
"URL": "http://dx.doi.org/10.1093/infdis/jiaf428",
"abstract": "<jats:title>Abstract</jats:title>\n <jats:p>Recent advances in infectious diseases diagnostics include the development, validation, and commercialization of new tests that measure host gene expression profiles or inflammatory protein concentrations. Interrogating host immune responses may help separate infectious from non-infectious inflammation, differentiate infection types and/or predict sepsis severity/subtype. This review summarizes the current state-of-the art in host response (HR) diagnostics for infectious diseases with a focus on test accuracy. Few studies have assessed the potential impact of HR testing on patient outcomes. We summarize current clinical evidence gaps and describe the types of studies needed to inform optimal integration in clinical practice.</jats:p>",
"article-number": "jiaf428",
"author": [
{
"ORCID": "https://orcid.org/0000-0001-9790-277X",
"affiliation": [
{
"name": "Department of Medicine, Infectious Diseases Division, University of Utah , Salt Lake City, UT ,",
"place": [
"USA"
]
},
{
"name": "Department of Pathology, Clinical Microbiology Section, University of Utah , Salt Lake City, UT ,",
"place": [
"USA"
]
},
{
"name": "ARUP Laboratories , Salt Lake City, UT ,",
"place": [
"USA"
]
}
],
"authenticated-orcid": false,
"family": "Hanson",
"given": "Kimberly E",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Danaher Diagnostics , Washington, DC ,",
"place": [
"USA"
]
},
{
"name": "Durham VA Health Care System , Durham, NC ,",
"place": [
"USA"
]
},
{
"name": "Duke University of School of Medicine , Durham, NC .",
"place": [
"USA"
]
}
],
"family": "Tsalik",
"given": "Ephraim L",
"sequence": "additional"
}
],
"container-title": "The Journal of Infectious Diseases",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
8,
8
]
],
"date-time": "2025-08-08T12:06:28Z",
"timestamp": 1754654788000
},
"deposited": {
"date-parts": [
[
2025,
8,
12
]
],
"date-time": "2025-08-12T23:55:38Z",
"timestamp": 1755042938000
},
"indexed": {
"date-parts": [
[
2025,
8,
15
]
],
"date-time": "2025-08-15T02:24:46Z",
"timestamp": 1755224686982,
"version": "3.43.0"
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2025,
8,
11
]
]
},
"language": "en",
"license": [
{
"URL": "https://academic.oup.com/pages/standard-publication-reuse-rights",
"content-version": "am",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
8,
11
]
],
"date-time": "2025-08-11T00:00:00Z",
"timestamp": 1754870400000
}
}
],
"link": [
{
"URL": "https://academic.oup.com/jid/advance-article-pdf/doi/10.1093/infdis/jiaf428/64017346/jiaf428.pdf",
"content-type": "application/pdf",
"content-version": "am",
"intended-application": "syndication"
},
{
"URL": "https://academic.oup.com/jid/advance-article-pdf/doi/10.1093/infdis/jiaf428/64017346/jiaf428.pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "286",
"original-title": [],
"prefix": "10.1093",
"published": {
"date-parts": [
[
2025,
8,
11
]
]
},
"published-online": {
"date-parts": [
[
2025,
8,
11
]
]
},
"publisher": "Oxford University Press (OUP)",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "https://academic.oup.com/jid/advance-article/doi/10.1093/infdis/jiaf428/8231658"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Host Immune Response Profiling for the Diagnosis of Infectious Diseases",
"type": "journal-article"
}