Prevalence of SARS-CoV-2 Omicron Sublineages and Spike Protein Mutations Conferring Resistance against Monoclonal Antibodies in a Swedish Cohort during 2022–2023
et al., Microorganisms, doi:10.3390/microorganisms11102417, Sep 2023
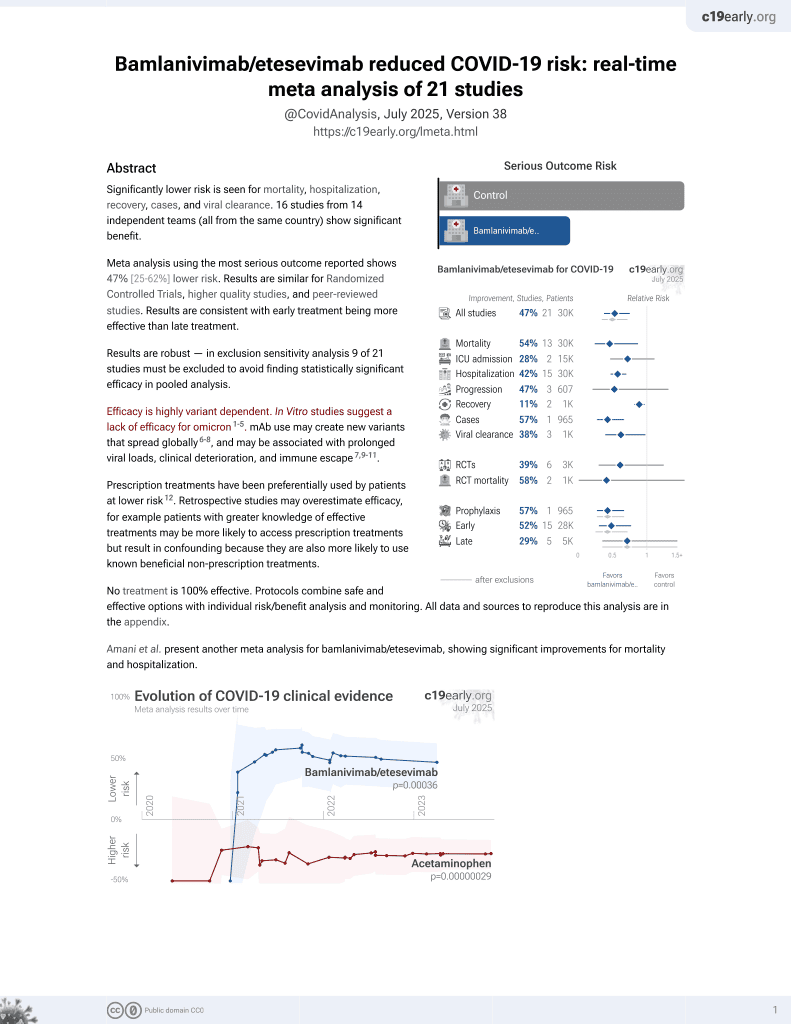
25th treatment shown to reduce risk in
May 2021, now with p = 0.00049 from 22 studies, recognized in 11 countries.
Efficacy is variant dependent.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
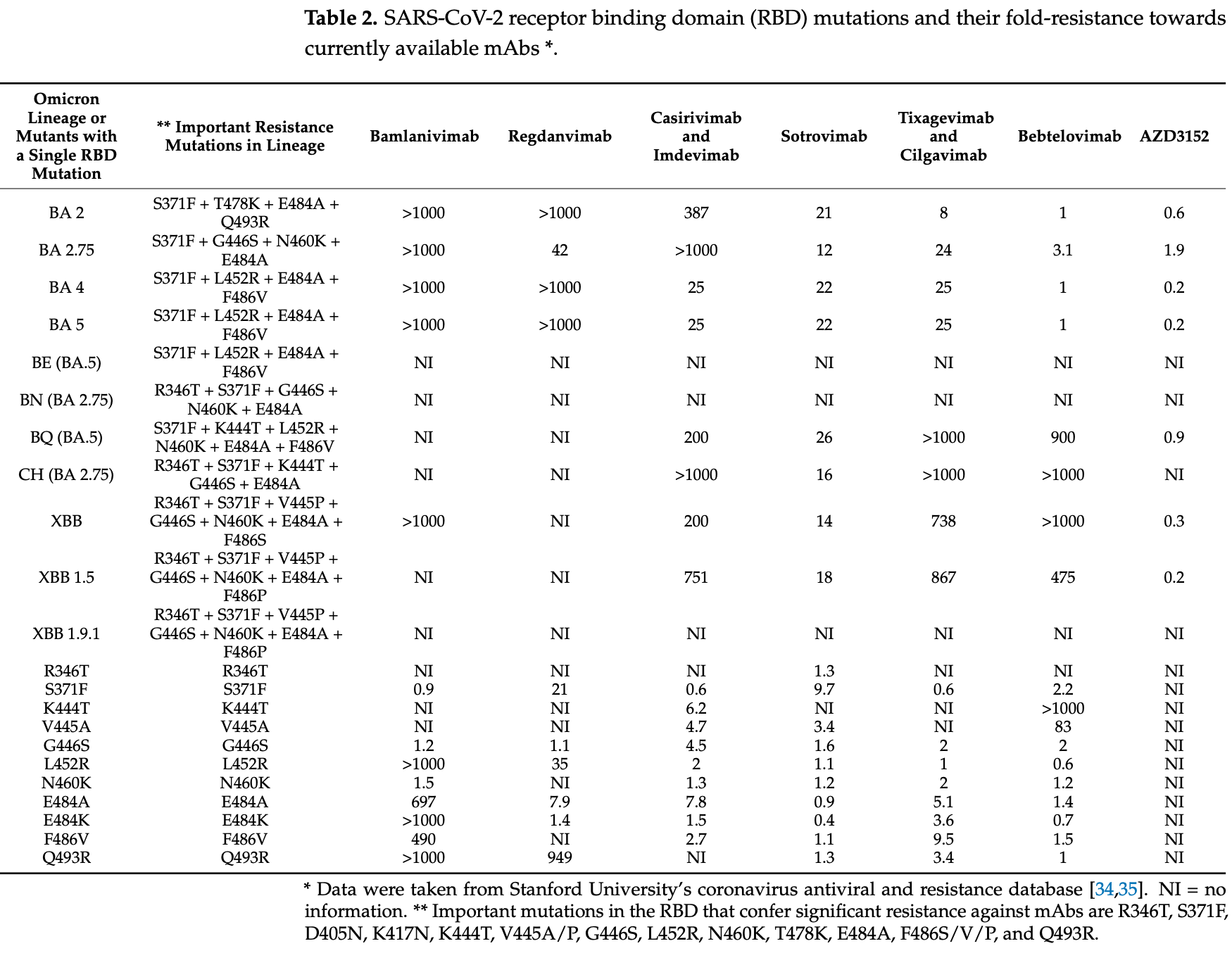
Analysis of 7,950 SARS-CoV-2 samples from central Sweden collected between March 2022 and May 2023 tracking the prevalence of omicron sublineages and mutations in the spike protein conferring resistance to monoclonal antibodies over time. Authors found the dominant sublineages shifted from BA.2 to BA.5 and its descendants during mid 2022, then to more diverse mix of BA.2 and BA.5 sublineages and their descendants towards end of 2022 and into 2023. Notably, mutations such as R346T, K444T, and V445P became increasingly common, rendering current monoclonal antibodies ineffective for most patients in the region. The findings highlight the continued need to monitor evolving SARS-CoV-2 mutations and develop new monoclonal antibodies targeting conserved spike protein regions.
Study covers bamlanivimab/etesevimab, regdanvimab, casirivimab/imdevimab, sotrovimab, tixagevimab/cilgavimab, and bebtelovimab.
1.
Liu et al., Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2, bioRxiv, doi:10.1101/2021.12.14.472719.
2.
Sheward et al., Variable loss of antibody potency against SARS-CoV-2 B.1.1.529 (Omicron), bioRxiv, doi:10.1101/2021.12.19.473354.
3.
VanBlargan et al., An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by several therapeutic monoclonal antibodies, bioRxiv, doi:10.1101/2021.12.15.472828.
Haars et al., 27 Sep 2023, Sweden, peer-reviewed, 9 authors.
Contact: johan.lennerstrand@medsci.uu.se (corresponding author), jonathan.haars@uu.se, johan.lindh@icm.uu.se, patrik.ellstrom@medsci.uu.se, rene.kaden@medsci.uu.se, n.palanisamy@chester.ac.uk, frans.wallin@regionorebrolan.se, paula.molling@oru.se, martin.sundqvist@oru.se.
Prevalence of SARS-CoV-2 Omicron Sublineages and Spike Protein Mutations Conferring Resistance against Monoclonal Antibodies in a Swedish Cohort during 2022–2023
Microorganisms, doi:10.3390/microorganisms11102417
Monoclonal antibodies (mAbs) are an important treatment option for COVID-19 caused by SARS-CoV-2, especially in immunosuppressed patients. However, this treatment option can become ineffective due to mutations in the SARS-CoV-2 genome, mainly in the receptor binding domain (RBD) of the spike (S) protein. In the present study, 7950 SARS-CoV-2 positive samples from the Uppsala and Örebro regions of central Sweden, collected between March 2022 and May 2023, were whole-genome sequenced using amplicon-based sequencing methods on Oxford Nanopore GridION, Illumina MiSeq, Illumina HiSeq, or MGI DNBSEQ-G400 instruments. Pango lineages were determined and all single nucleotide polymorphism (SNP) mutations that occurred in these samples were identified. We found that the dominant sublineages changed over time, and mutations conferring resistance to currently available mAbs became common. Notable ones are R346T and K444T mutations in the RBD that confer significant resistance against tixagevimab and cilgavimab mAbs. Further, mutations conferring a high-fold resistance to bebtelovimab, such as the K444T and V445P mutations, were also observed in the samples. This study highlights that resistance mutations have over time rendered currently available mAbs ineffective against SARS-CoV-2 in most patients. Therefore, there is a need for continued surveillance of resistance mutations and the development of new mAbs that target more conserved regions of the RBD.
The repeated events of evolved resistance to mAbs means that the development of new mAbs and other antiviral treatments remains important for patients infected with SARS-CoV-2. The rapid increase in resistance to tixagevimab and cilgavimab highlights that these changes need to be communicated quickly by labs and scientists to the public. Therefore, the continued surveillance of SARS-CoV-2 through whole-genome sequencing is essential for understanding the evolution of the virus and for providing scientists, physicians, patients, decision makers, and drug manufacturers with correct and updated information for curbing this infection in the population.
Supplementary Materials: The following supporting information can be downloaded at: https: //www.mdpi.com/article/10.3390/microorganisms11102417/s1, Figure S1 Author Contributions: Conceptualization, J.H., N.P. and J.L. (Johan Lennerstrand); methodology, J.H. and J.L. (Johan Lennerstrand); software, J.H. and R.K.; formal analysis, J.H.; investigation, J.H., F.W., P.M., P.E., R.K. and J.L. (Johan Lennerstrand); resources, J.L. (Johan Lindh), M.S. and R.K.; data curation, J.H. and F.W.; writing-original draft preparation, J.H.; writing-review and editing, J.H., N.P., F.W., P.M., P.E., J.L. (Johan Lindh), M.S., R.K. and J.L. (Johan Lennerstrand); visualization, J.H.; supervision, R.K. and J.L. (Johan Lennerstrand); funding acquisition, J.L. (Johan Lennerstrand). All authors have read and agreed to the published version of the..
References
Astrazeneca, A Phase I/III Randomized, Double Blind Study to Evaluate the Safety, Efficacy and Neutralizing Activity of AZD5156/AZD3152 for Pre Exposure Prophylaxis of COVID 19 in Participants With Conditions Causing Immune Impairment. Sub-Study: Phase II Open Label Sub-Study to Evaluate the Safety, PK, and Neutralizing Activity of AZD3152 for Pre-Exposure Prophylaxis of COVID-19
Barnes, Jette, Abernathy, Dam, Esswein et al., SARS-CoV-2 Neutralizing Antibody Structures Inform Therapeutic Strategies, Nature, doi:10.1038/s41586-020-2852-1
Bbmap, None
Cao, Jian, Wang, Yu, Song et al., Imprinted SARS-CoV-2 Humoral Immunity Induces Convergent Omicron RBD Evolution, Nature, doi:10.1038/s41586-022-05644-7
Casadevall, Focosi, SARS-CoV-2 Variants Resistant to Monoclonal Antibodies in Immunocompromised Patients Constitute a Public Health Concern, J. Clin. Investig, doi:10.1172/JCI168603
Chen, Zhou, Chen, Gu, Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor, Bioinformatics, doi:10.1093/bioinformatics/bty560
Davis, Long, Christensen, Olsen, Olson et al., Analysis of the ARTIC Version 3 and Version 4 SARS-CoV-2 Primers and Their Impact on the Detection of the G142D Amino Acid Substitution in the Spike Protein, Microbiol. Spectr, doi:10.1128/Spectrum.01803-21
Ema, EMA Issues Advice on Use of Antibody Combination (Bamlanivimab/Etesevimab)
Evusheld, None
Focosi, Novazzi, Genoni, Dentali, Gasperina et al., Emergence of SARS-COV-2 Spike Protein Escape Mutation Q493R after Treatment for COVID-19, Emerg. Infect. Dis, doi:10.3201/eid2710.211538
Freed, Vlková, Faisal, Silander, Rapid and Inexpensive Whole-Genome Sequencing of SARS-CoV-2 Using 1200 Bp Tiled Amplicons and Oxford Nanopore Rapid Barcoding, Biol. Methods Protoc, doi:10.1093/biomethods/bpaa014
Garrison, Marth, Haplotype-Based Variant Detection from Short-Read Sequencing
Gisaid-Gisaid, None
Gms-Artic, None
Grubaugh, Gangavarapu, Quick, Matteson, De Jesus et al., An Amplicon-Based Sequencing Framework for Accurately Measuring Intrahost Virus Diversity Using PrimalSeq and iVar, Genome Biol, doi:10.1186/s13059-018-1618-7
Gupta, Konnova, Smet, Berkell, Savoldi et al., Host Immunological Responses Facilitate Development of SARS-CoV-2 Mutations in Patients Receiving Monoclonal Antibody Treatments, J. Clin. Investig, doi:10.1172/JCI166032
Hoffmann, Wong, Arora, Zhang, Rocha et al., BA.5 Efficiently Infects Lung Cells, Nat. Commun, doi:10.1038/s41467-023-39147-4
Iketani, Liu, Guo, Liu, Chan et al., Antibody Evasion Properties of SARS-CoV-2 Omicron Sublineages, Nature, doi:10.1038/s41586-022-04594-4
Jünemann, Sedlazeck, Prior, Albersmeier, John et al., Updating Benchtop Sequencing Performance Comparison, Nat. Biotechnol, doi:10.1038/nbt.2522
Kaden, Early Phylogenetic Diversification of SARS-CoV-2: Determination of Variants and the Effect on Epidemiology, Immunology, and Diagnostics, J. Clin. Med, doi:10.3390/jcm9061615
Lennerstrand, Palanisamy, Global Prevalence of Adaptive and Prolonged Infections' Mutations in the Receptor-Binding Domain of the SARS-CoV-2 Spike Protein, Viruses, doi:10.3390/v13101974
Li, Minimap2: Pairwise Alignment for Nucleotide Sequences, Bioinformatics, doi:10.1093/bioinformatics/bty191
Lyke, Atmar, Islas, Posavad, Szydlo et al., Rapid Decline in Vaccine-Boosted Neutralizing Antibodies against SARS-CoV-2 Omicron Variant, Cell Rep. Med, doi:10.1016/j.xcrm.2022.100679
Mannsverk, Bergholm, Palanisamy, Ellström, Kaden et al., SARS-CoV-2 Variants of Concern and Spike Protein Mutational Dynamics in a Swedish Cohort during 2021, Studied by Nanopore Sequencing, Virol. J, doi:10.1186/s12985-022-01896-x
Mccormick, Jacobs, Mellors, The Emerging Plasticity of SARS-CoV-2, Science, doi:10.1126/science.abg4493
Meng, Abdullahi, Ferreira, Goonawardane, Saito et al., Altered TMPRSS2 Usage by SARS-CoV-2 Omicron Impacts Infectivity and Fusogenicity, Nature, doi:10.1038/s41586-022-04474-x
Mercatelli, Triboli, Fornasari, Ray, Giorgi, Coronapp: A Web Application to Annotate and Monitor SARS-CoV-2 Mutations, J. Med. Virol, doi:10.1002/jmv.26678
O'toole, Scher, Underwood, Jackson, Hill et al., Assignment of Epidemiological Lineages in an Emerging Pandemic Using the Pangolin Tool, Virus Evol, doi:10.1093/ve/veab064
Owen, Allerton, Anderson, Aschenbrenner, Avery et al., An Oral SARS-CoV-2 Mpro Inhibitor Clinical Candidate for the Treatment of COVID-19, Science, doi:10.1126/science.abl4784
Pillay, Gene of the Month: The 2019-nCoV/SARS-CoV-2 Novel Coronavirus Spike Protein, J. Clin. Pathol, doi:10.1136/jclinpath-2020-206658
Quick, Sequencing Protocol v3 (LoCost)
Rambaut, Holmes, O'toole, Hill, Mccrone et al., A Dynamic Nomenclature Proposal for SARS-CoV-2 Lineages to Assist Genomic Epidemiology, Nat. Microbiol, doi:10.1038/s41564-020-0770-5
Regkirona, None
Schmutz, Pango Lineage Translator
Shrestha, Tedla, Bull, Broadly-Neutralizing Antibodies Against Emerging SARS-CoV-2 Variants, Front. Immunol, doi:10.3389/fimmu.2021.752003
Starr, Greaney, Dingens, Bloom, Complete Map of SARS-CoV-2 RBD Mutations That Escape the Monoclonal Antibody LY-CoV555 and Its Cocktail with LY-CoV016, Cell Rep. Med, doi:10.1016/j.xcrm.2021.100255
Sydow, Lindqvist, Asghar, Johansson, Sundqvist et al., Comparison of SARS-CoV-2 Whole Genome Sequencing Using Tiled Amplicon Enrichment and Bait Hybridization, Sci. Rep, doi:10.1038/s41598-023-33168-1
Tai, He, Zhang, Pu, Voronin et al., Characterization of the Receptor-Binding Domain (RBD) of 2019 Novel Coronavirus: Implication for Development of RBD Protein as a Viral Attachment Inhibitor and Vaccine, Cell Mol. Immunol, doi:10.1038/s41423-020-0400-4
Tamura, Ito, Uriu, Zahradnik, Kida et al., Virological Characteristics of the SARS-CoV-2 XBB Variant Derived from Recombination of Two Omicron Subvariants, Nat. Commun, doi:10.1038/s41467-023-38435-3
Truffot, Andréani, Le Maréchal, Caporossi, Epaulard et al., SARS-CoV-2 Variants in Immunocompromised Patient Given Antibody Monotherapy, Emerg. Infect. Dis, doi:10.3201/eid2710.211509
Tyson, James, Stoddart, Sparks, Wickenhagen et al., Improvements to the ARTIC Multiplex PCR Method for SARS-CoV-2 Genome Sequencing Using Nanopore, bioRxiv, doi:10.1101/2020.09.04.283077
Töpfer, Cbg-Ethz, /ConsensusFixer: Computes a Consensus Sequence with Wobbles, Ambiguous Bases, and in-Frame Insertions, from a NGS Read Alignment
Vellas, Trémeaux, Bello, Latour, Jeanne et al., Resistance Mutations in SARS-CoV-2 Omicron Variant in Patients Treated with Sotrovimab, Clin. Microbiol. Infect, doi:10.1016/j.cmi.2022.05.002
Viana, Moyo, Amoako, Tegally, Scheepers et al., Rapid Epidemic Expansion of the SARS-CoV-2 Omicron Variant in Southern Africa, Nature, doi:10.1038/s41586-022-04411-y
Wang, Iketani, Li, Liu, Guo et al., Alarming Antibody Evasion Properties of Rising SARS-CoV-2 BQ and XBB Subvariants, Cell, doi:10.1016/j.cell.2022.12.018
Yue, Song, Wang, Jian, Chen et al., ACE2 Binding and Antibody Evasion in Enhanced Transmissibility of XBB.1.5, Lancet Infect. Dis, doi:10.1016/S1473-3099(23)00010-5
Zhao, Lu, Peng, Chen, Meng et al., SARS-CoV-2 Omicron Variant Shows Less Efficient Replication and Fusion Activity When Compared with Delta Variant in TMPRSS2-Expressed Cells, Emerg. Microbes Infect, doi:10.1080/22221751.2021.2023329
DOI record:
{
"DOI": "10.3390/microorganisms11102417",
"ISSN": [
"2076-2607"
],
"URL": "http://dx.doi.org/10.3390/microorganisms11102417",
"abstract": "<jats:p>Monoclonal antibodies (mAbs) are an important treatment option for COVID-19 caused by SARS-CoV-2, especially in immunosuppressed patients. However, this treatment option can become ineffective due to mutations in the SARS-CoV-2 genome, mainly in the receptor binding domain (RBD) of the spike (S) protein. In the present study, 7950 SARS-CoV-2 positive samples from the Uppsala and Örebro regions of central Sweden, collected between March 2022 and May 2023, were whole-genome sequenced using amplicon-based sequencing methods on Oxford Nanopore GridION, Illumina MiSeq, Illumina HiSeq, or MGI DNBSEQ-G400 instruments. Pango lineages were determined and all single nucleotide polymorphism (SNP) mutations that occurred in these samples were identified. We found that the dominant sublineages changed over time, and mutations conferring resistance to currently available mAbs became common. Notable ones are R346T and K444T mutations in the RBD that confer significant resistance against tixagevimab and cilgavimab mAbs. Further, mutations conferring a high-fold resistance to bebtelovimab, such as the K444T and V445P mutations, were also observed in the samples. This study highlights that resistance mutations have over time rendered currently available mAbs ineffective against SARS-CoV-2 in most patients. Therefore, there is a need for continued surveillance of resistance mutations and the development of new mAbs that target more conserved regions of the RBD.</jats:p>",
"alternative-id": [
"microorganisms11102417"
],
"author": [
{
"ORCID": "http://orcid.org/0009-0003-8735-4097",
"affiliation": [
{
"name": "Department of Medical Sciences, Section for Clinical Microbiology and Hospital Hygiene Uppsala University, Akademiska Sjukhuset Entrance 40 Floor 5, 751 85 Uppsala, Sweden"
}
],
"authenticated-orcid": false,
"family": "Haars",
"given": "Jonathan",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0003-0369-2316",
"affiliation": [
{
"name": "Chester Medical School, University of Chester, Chester CH2 1BR, UK"
}
],
"authenticated-orcid": false,
"family": "Palanisamy",
"given": "Navaneethan",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Laboratory Medicine, Clinical Microbiology, Örebro University Hospital, Södra Grev Rosengatan, 701 85 Örebro, Sweden"
}
],
"family": "Wallin",
"given": "Frans",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Laboratory Medicine, Clinical Microbiology, Faculty of Medicine and Health, Örebro University, 701 82 Örebro, Sweden"
}
],
"family": "Mölling",
"given": "Paula",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Medical Sciences, Section for Clinical Microbiology and Hospital Hygiene Uppsala University, Akademiska Sjukhuset Entrance 40 Floor 5, 751 85 Uppsala, Sweden"
}
],
"family": "Lindh",
"given": "Johan",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Laboratory Medicine, Clinical Microbiology, Faculty of Medicine and Health, Örebro University, 701 82 Örebro, Sweden"
}
],
"family": "Sundqvist",
"given": "Martin",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Medical Sciences, Section for Clinical Microbiology and Hospital Hygiene Uppsala University, Akademiska Sjukhuset Entrance 40 Floor 5, 751 85 Uppsala, Sweden"
}
],
"family": "Ellström",
"given": "Patrik",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-2111-9751",
"affiliation": [
{
"name": "Department of Medical Sciences, Section for Clinical Microbiology and Hospital Hygiene Uppsala University, Akademiska Sjukhuset Entrance 40 Floor 5, 751 85 Uppsala, Sweden"
},
{
"name": "SciLifeLab, Clinical Genomics Uppsala, Husargatan 3, 752 37 Uppsala, Sweden"
}
],
"authenticated-orcid": false,
"family": "Kaden",
"given": "René",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Medical Sciences, Section for Clinical Microbiology and Hospital Hygiene Uppsala University, Akademiska Sjukhuset Entrance 40 Floor 5, 751 85 Uppsala, Sweden"
}
],
"family": "Lennerstrand",
"given": "Johan",
"sequence": "additional"
}
],
"container-title": "Microorganisms",
"container-title-short": "Microorganisms",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2023,
9,
28
]
],
"date-time": "2023-09-28T05:42:12Z",
"timestamp": 1695879732000
},
"deposited": {
"date-parts": [
[
2023,
9,
28
]
],
"date-time": "2023-09-28T07:10:47Z",
"timestamp": 1695885047000
},
"funder": [
{
"award": [
"RFR-980115"
],
"name": "Regional Research Council Mid Sweden"
}
],
"indexed": {
"date-parts": [
[
2023,
9,
29
]
],
"date-time": "2023-09-29T08:46:48Z",
"timestamp": 1695977208200
},
"is-referenced-by-count": 0,
"issue": "10",
"issued": {
"date-parts": [
[
2023,
9,
27
]
]
},
"journal-issue": {
"issue": "10",
"published-online": {
"date-parts": [
[
2023,
10
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
9,
27
]
],
"date-time": "2023-09-27T00:00:00Z",
"timestamp": 1695772800000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/2076-2607/11/10/2417/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "2417",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2023,
9,
27
]
]
},
"published-online": {
"date-parts": [
[
2023,
9,
27
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"key": "ref_1",
"unstructured": "(2023, August 22). IHR Emergency Committee on Novel Coronavirus (2019-nCoV). Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-statement-on-ihr-emergency-committee-on-novel-coronavirus-(2019-ncov)."
},
{
"DOI": "10.1016/j.xcrm.2022.100679",
"article-title": "Rapid Decline in Vaccine-Boosted Neutralizing Antibodies against SARS-CoV-2 Omicron Variant",
"author": "Lyke",
"doi-asserted-by": "crossref",
"first-page": "100679",
"journal-title": "Cell Rep. Med.",
"key": "ref_2",
"volume": "3",
"year": "2022"
},
{
"DOI": "10.1126/science.abl4784",
"article-title": "An Oral SARS-CoV-2 Mpro Inhibitor Clinical Candidate for the Treatment of COVID-19",
"author": "Owen",
"doi-asserted-by": "crossref",
"first-page": "1586",
"journal-title": "Science",
"key": "ref_3",
"volume": "374",
"year": "2021"
},
{
"DOI": "10.3389/fimmu.2021.752003",
"article-title": "Broadly-Neutralizing Antibodies Against Emerging SARS-CoV-2 Variants",
"author": "Shrestha",
"doi-asserted-by": "crossref",
"first-page": "4025",
"journal-title": "Front. Immunol.",
"key": "ref_4",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1038/s41586-020-2852-1",
"article-title": "SARS-CoV-2 Neutralizing Antibody Structures Inform Therapeutic Strategies",
"author": "Barnes",
"doi-asserted-by": "crossref",
"first-page": "682",
"journal-title": "Nature",
"key": "ref_5",
"volume": "588",
"year": "2020"
},
{
"DOI": "10.1136/jclinpath-2020-206658",
"article-title": "Gene of the Month: The 2019-nCoV/SARS-CoV-2 Novel Coronavirus Spike Protein",
"author": "Pillay",
"doi-asserted-by": "crossref",
"first-page": "366",
"journal-title": "J. Clin. Pathol.",
"key": "ref_6",
"volume": "73",
"year": "2020"
},
{
"DOI": "10.1038/s41423-020-0400-4",
"article-title": "Characterization of the Receptor-Binding Domain (RBD) of 2019 Novel Coronavirus: Implication for Development of RBD Protein as a Viral Attachment Inhibitor and Vaccine",
"author": "Tai",
"doi-asserted-by": "crossref",
"first-page": "613",
"journal-title": "Cell Mol. Immunol.",
"key": "ref_7",
"volume": "17",
"year": "2020"
},
{
"key": "ref_8",
"unstructured": "USFDA (2023, August 22). Coronavirus (COVID-19) Update: FDA Revokes Emergency Use Authorization for Monoclonal Antibody Bamlanivimab, Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-revokes-emergency-use-authorization-monoclonal-antibody-bamlanivimab."
},
{
"key": "ref_9",
"unstructured": "EMA (2023, August 22). EMA Issues Advice on Use of Antibody Combination (Bamlanivimab/Etesevimab). Available online: https://www.ema.europa.eu/en/news/ema-issues-advice-use-antibody-combination-bamlanivimab-etesevimab."
},
{
"key": "ref_10",
"unstructured": "EMA (2023, August 22). Bamlanivimab and Etesevimab for COVID-19: Withdrawn Application. Available online: https://www.ema.europa.eu/en/medicines/human/withdrawn-applications/bamlanivimab-etesevimab-covid-19."
},
{
"key": "ref_11",
"unstructured": "(2023, August 22). Emergency Use Authorization 094. Available online: https://pi.lilly.com/eua/bam-and-ete-eua-fda-authorization-letter.pdf."
},
{
"key": "ref_12",
"unstructured": "EMA (2023, August 22). Regkirona. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/regkirona."
},
{
"key": "ref_13",
"unstructured": "USFDA (2023, August 22). Coronavirus (COVID-19) Update: FDA Limits Use of Certain Monoclonal Antibodies to Treat COVID-19 Due to the Omicron Variant, Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-limits-use-certain-monoclonal-antibodies-treat-covid-19-due-omicron."
},
{
"key": "ref_14",
"unstructured": "USFDA (2023, August 22). Coronavirus (COVID-19) Update: FDA Authorizes Additional Monoclonal Antibody for Treatment of COVID-19, Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-additional-monoclonal-antibody-treatment-covid-19."
},
{
"key": "ref_15",
"unstructured": "USFDA (2023, August 22). FDA Roundup: 5 April 2022, Available online: https://www.fda.gov/news-events/press-announcements/fda-roundup-april-5-2022."
},
{
"key": "ref_16",
"unstructured": "EMA (2023, August 22). Xevudy. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/xevudy."
},
{
"key": "ref_17",
"unstructured": "USFDA (2023, August 22). Coronavirus (COVID-19) Update: FDA Authorizes New Long-Acting Monoclonal Antibodies for Pre-Exposure Prevention of COVID-19 in Certain Individuals, Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-new-long-acting-monoclonal-antibodies-pre-exposure."
},
{
"key": "ref_18",
"unstructured": "USFDA (2023, August 22). Announces Evusheld Is Not Currently Authorized for Emergency Use in the U.S, Available online: https://www.fda.gov/drugs/drug-safety-and-availability/fda-announces-evusheld-not-currently-authorized-emergency-use-us."
},
{
"key": "ref_19",
"unstructured": "EMA (2023, August 22). Evusheld. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/evusheld."
},
{
"key": "ref_20",
"unstructured": "USFDA (2023, August 22). Coronavirus (COVID-19) Update: FDA Authorizes New Monoclonal Antibody for Treatment of COVID-19 That Retains Activity Against Omicron Variant, Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-new-monoclonal-antibody-treatment-covid-19-retains."
},
{
"key": "ref_21",
"unstructured": "USFDA (2023, August 22). Announces Bebtelovimab Is Not Currently Authorized in Any US Region, Available online: https://www.fda.gov/drugs/drug-safety-and-availability/fda-announces-bebtelovimab-not-currently-authorized-any-us-region."
},
{
"DOI": "10.3390/jcm9061615",
"doi-asserted-by": "crossref",
"key": "ref_22",
"unstructured": "Kaden, R. (2020). Early Phylogenetic Diversification of SARS-CoV-2: Determination of Variants and the Effect on Epidemiology, Immunology, and Diagnostics. J. Clin. Med., 9."
},
{
"DOI": "10.1126/science.abg4493",
"article-title": "The Emerging Plasticity of SARS-CoV-2",
"author": "McCormick",
"doi-asserted-by": "crossref",
"first-page": "1306",
"journal-title": "Science",
"key": "ref_23",
"volume": "371",
"year": "2021"
},
{
"DOI": "10.3390/v13101974",
"doi-asserted-by": "crossref",
"key": "ref_24",
"unstructured": "Lennerstrand, J., and Palanisamy, N. (2021). Global Prevalence of Adaptive and Prolonged Infections’ Mutations in the Receptor-Binding Domain of the SARS-CoV-2 Spike Protein. Viruses, 13."
},
{
"DOI": "10.1016/j.cell.2022.12.018",
"article-title": "Alarming Antibody Evasion Properties of Rising SARS-CoV-2 BQ and XBB Subvariants",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "279",
"journal-title": "Cell",
"key": "ref_25",
"volume": "186",
"year": "2023"
},
{
"article-title": "Imprinted SARS-CoV-2 Humoral Immunity Induces Convergent Omicron RBD Evolution",
"author": "Cao",
"first-page": "521",
"journal-title": "Nature",
"key": "ref_26",
"volume": "614",
"year": "2023"
},
{
"DOI": "10.1038/s41586-022-04594-4",
"article-title": "Antibody Evasion Properties of SARS-CoV-2 Omicron Sublineages",
"author": "Iketani",
"doi-asserted-by": "crossref",
"first-page": "553",
"journal-title": "Nature",
"key": "ref_27",
"volume": "604",
"year": "2022"
},
{
"DOI": "10.1016/j.xcrm.2021.100255",
"article-title": "Complete Map of SARS-CoV-2 RBD Mutations That Escape the Monoclonal Antibody LY-CoV555 and Its Cocktail with LY-CoV016",
"author": "Starr",
"doi-asserted-by": "crossref",
"first-page": "100255",
"journal-title": "Cell Rep. Med.",
"key": "ref_28",
"volume": "2",
"year": "2021"
},
{
"DOI": "10.3201/eid2710.211538",
"article-title": "Emergence of SARS-COV-2 Spike Protein Escape Mutation Q493R after Treatment for COVID-19",
"author": "Focosi",
"doi-asserted-by": "crossref",
"first-page": "2728",
"journal-title": "Emerg. Infect. Dis.",
"key": "ref_29",
"volume": "27",
"year": "2021"
},
{
"DOI": "10.3201/eid2710.211509",
"article-title": "SARS-CoV-2 Variants in Immunocompromised Patient Given Antibody Monotherapy",
"author": "Truffot",
"doi-asserted-by": "crossref",
"first-page": "2725",
"journal-title": "Emerg. Infect. Dis.",
"key": "ref_30",
"volume": "27",
"year": "2021"
},
{
"DOI": "10.1172/JCI168603",
"doi-asserted-by": "crossref",
"key": "ref_31",
"unstructured": "Casadevall, A., and Focosi, D. (2023). SARS-CoV-2 Variants Resistant to Monoclonal Antibodies in Immunocompromised Patients Constitute a Public Health Concern. J. Clin. Investig., 133."
},
{
"DOI": "10.1172/JCI166032",
"doi-asserted-by": "crossref",
"key": "ref_32",
"unstructured": "Gupta, A., Konnova, A., Smet, M., Berkell, M., Savoldi, A., Morra, M., Averbeke, V.V., Winter, F.H.R.D., Peserico, D., and Danese, E. (2023). Host Immunological Responses Facilitate Development of SARS-CoV-2 Mutations in Patients Receiving Monoclonal Antibody Treatments. J. Clin. Investig., 133."
},
{
"DOI": "10.1016/j.cmi.2022.05.002",
"article-title": "Resistance Mutations in SARS-CoV-2 Omicron Variant in Patients Treated with Sotrovimab",
"author": "Vellas",
"doi-asserted-by": "crossref",
"first-page": "1297",
"journal-title": "Clin. Microbiol. Infect.",
"key": "ref_33",
"volume": "28",
"year": "2022"
},
{
"DOI": "10.1038/s41586-022-04411-y",
"article-title": "Rapid Epidemic Expansion of the SARS-CoV-2 Omicron Variant in Southern Africa",
"author": "Viana",
"doi-asserted-by": "crossref",
"first-page": "679",
"journal-title": "Nature",
"key": "ref_34",
"volume": "603",
"year": "2022"
},
{
"DOI": "10.1038/s41586-022-04474-x",
"article-title": "Altered TMPRSS2 Usage by SARS-CoV-2 Omicron Impacts Infectivity and Fusogenicity",
"author": "Meng",
"doi-asserted-by": "crossref",
"first-page": "706",
"journal-title": "Nature",
"key": "ref_35",
"volume": "603",
"year": "2022"
},
{
"DOI": "10.1080/22221751.2021.2023329",
"article-title": "SARS-CoV-2 Omicron Variant Shows Less Efficient Replication and Fusion Activity When Compared with Delta Variant in TMPRSS2-Expressed Cells",
"author": "Zhao",
"doi-asserted-by": "crossref",
"first-page": "277",
"journal-title": "Emerg. Microbes Infect.",
"key": "ref_36",
"volume": "11",
"year": "2022"
},
{
"DOI": "10.1038/s41467-023-39147-4",
"article-title": "Omicron Subvariant BA.5 Efficiently Infects Lung Cells",
"author": "Hoffmann",
"doi-asserted-by": "crossref",
"first-page": "3500",
"journal-title": "Nat. Commun.",
"key": "ref_37",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1186/s12985-022-01896-x",
"article-title": "SARS-CoV-2 Variants of Concern and Spike Protein Mutational Dynamics in a Swedish Cohort during 2021, Studied by Nanopore Sequencing",
"author": "Mannsverk",
"doi-asserted-by": "crossref",
"first-page": "164",
"journal-title": "Virol. J.",
"key": "ref_38",
"volume": "19",
"year": "2022"
},
{
"DOI": "10.1038/s41598-023-33168-1",
"article-title": "Comparison of SARS-CoV-2 Whole Genome Sequencing Using Tiled Amplicon Enrichment and Bait Hybridization",
"author": "Lindqvist",
"doi-asserted-by": "crossref",
"first-page": "6461",
"journal-title": "Sci. Rep.",
"key": "ref_39",
"volume": "13",
"year": "2023"
},
{
"DOI": "10.1038/nbt.2522",
"article-title": "Updating Benchtop Sequencing Performance Comparison",
"author": "Sedlazeck",
"doi-asserted-by": "crossref",
"first-page": "294",
"journal-title": "Nat. Biotechnol.",
"key": "ref_40",
"volume": "31",
"year": "2013"
},
{
"key": "ref_41",
"unstructured": "(2023, September 08). Gms-Artic. Available online: https://github.com/genomic-medicine-sweden/gms-artic."
},
{
"key": "ref_42",
"unstructured": "(2023, September 08). Epi2me-Labs/Wf-Artic: ARTIC SARS-CoV-2 Workflow and Reporting. Available online: https://github.com/epi2me-labs/wf-artic."
},
{
"key": "ref_43",
"unstructured": "(2023, September 08). National Sample Collection Stored at NPC—The COVID-19 Library|Karolinska Institutet. Available online: https://ki.se/en/mtc/national-sample-collection-stored-at-npc-the-covid-19-library."
},
{
"DOI": "10.1093/bioinformatics/bty560",
"article-title": "Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor",
"author": "Chen",
"doi-asserted-by": "crossref",
"first-page": "i884",
"journal-title": "Bioinformatics",
"key": "ref_44",
"volume": "34",
"year": "2018"
},
{
"DOI": "10.1186/s13059-018-1618-7",
"doi-asserted-by": "crossref",
"key": "ref_45",
"unstructured": "Grubaugh, N.D., Gangavarapu, K., Quick, J., Matteson, N.L., De Jesus, J.G., Main, B.J., Tan, A.L., Paul, L.M., Brackney, D.E., and Grewal, S. (2019). An Amplicon-Based Sequencing Framework for Accurately Measuring Intrahost Virus Diversity Using PrimalSeq and iVar. Genome Biol., 20."
},
{
"key": "ref_46",
"unstructured": "Garrison, E., and Marth, G. (2012). Haplotype-Based Variant Detection from Short-Read Sequencing. arXiv."
},
{
"key": "ref_47",
"unstructured": "Töpfer, A. (2023, September 08). Cbg-Ethz/ConsensusFixer: Computes a Consensus Sequence with Wobbles, Ambiguous Bases, and in-Frame Insertions, from a NGS Read Alignment. Available online: https://github.com/cbg-ethz/consensusfixer."
},
{
"DOI": "10.1093/biomethods/bpaa014",
"doi-asserted-by": "crossref",
"key": "ref_48",
"unstructured": "Freed, N.E., Vlková, M., Faisal, M.B., and Silander, O.K. (2020). Rapid and Inexpensive Whole-Genome Sequencing of SARS-CoV-2 Using 1200 Bp Tiled Amplicons and Oxford Nanopore Rapid Barcoding. Biol. Methods Protoc., 5."
},
{
"DOI": "10.1101/2020.09.04.283077",
"doi-asserted-by": "crossref",
"key": "ref_49",
"unstructured": "Tyson, J.R., James, P., Stoddart, D., Sparks, N., Wickenhagen, A., Hall, G., Choi, J.H., Lapointe, H., Kamelian, K., and Smith, A.D. (2020). Improvements to the ARTIC Multiplex PCR Method for SARS-CoV-2 Genome Sequencing Using Nanopore. bioRxiv."
},
{
"DOI": "10.17504/protocols.io.bp2l6n26rgqe/v3",
"doi-asserted-by": "crossref",
"key": "ref_50",
"unstructured": "Quick, J. (2023, May 02). nCoV-2019 Sequencing Protocol v3 (LoCost). Available online: https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye."
},
{
"key": "ref_51",
"unstructured": "(2023, August 22). Can Coverage Be Improved for the BA.2 Variant?|NEB. Available online: https://international.neb.com/faqs/2022/04/28/can-coverage-be-improved-for-the-ba2-variant."
},
{
"DOI": "10.1128/Spectrum.01803-21",
"article-title": "Analysis of the ARTIC Version 3 and Version 4 SARS-CoV-2 Primers and Their Impact on the Detection of the G142D Amino Acid Substitution in the Spike Protein",
"author": "Davis",
"doi-asserted-by": "crossref",
"first-page": "e01803-21",
"journal-title": "Microbiol. Spectr.",
"key": "ref_52",
"volume": "9",
"year": "2021"
},
{
"key": "ref_53",
"unstructured": "(2023, August 22). Geneious|Bioinformatics Software for Sequence Data Analysis. Available online: https://www.geneious.com/."
},
{
"key": "ref_54",
"unstructured": "(2023, August 22). BBMap. Available online: https://sourceforge.net/projects/bbmap/."
},
{
"DOI": "10.1093/bioinformatics/bty191",
"article-title": "Minimap2: Pairwise Alignment for Nucleotide Sequences",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "3094",
"journal-title": "Bioinformatics",
"key": "ref_55",
"volume": "34",
"year": "2018"
},
{
"key": "ref_56",
"unstructured": "(2023, September 20). Sequencing of SARS-CoV-2—First Update. Available online: https://www.ecdc.europa.eu/en/publications-data/sequencing-sars-cov-2."
},
{
"key": "ref_57",
"unstructured": "(2023, August 22). GISAID—Gisaid.Org. Available online: https://gisaid.org/."
},
{
"DOI": "10.1038/s41564-020-0770-5",
"article-title": "A Dynamic Nomenclature Proposal for SARS-CoV-2 Lineages to Assist Genomic Epidemiology",
"author": "Rambaut",
"doi-asserted-by": "crossref",
"first-page": "1403",
"journal-title": "Nat. Microbiol.",
"key": "ref_58",
"volume": "5",
"year": "2020"
},
{
"key": "ref_59",
"unstructured": "(2023, August 29). Geneious Wrapper Plugin for Pangolin. Available online: https://github.com/clinical-genomics-uppsala/Geneious_pangolin_wrapper."
},
{
"DOI": "10.1093/ve/veab064",
"article-title": "Assignment of Epidemiological Lineages in an Emerging Pandemic Using the Pangolin Tool",
"author": "Scher",
"doi-asserted-by": "crossref",
"first-page": "veab064",
"journal-title": "Virus Evol.",
"key": "ref_60",
"volume": "7",
"year": "2021"
},
{
"key": "ref_61",
"unstructured": "Schmutz, S. (2023, August 22). Pango Lineage Translator. Available online: https://github.com/sschmutz/PangoLineageTranslator."
},
{
"DOI": "10.1002/jmv.26678",
"article-title": "Coronapp: A Web Application to Annotate and Monitor SARS-CoV-2 Mutations",
"author": "Mercatelli",
"doi-asserted-by": "crossref",
"first-page": "3238",
"journal-title": "J. Med. Virol.",
"key": "ref_62",
"volume": "93",
"year": "2021"
},
{
"DOI": "10.1038/s41467-023-38435-3",
"article-title": "Virological Characteristics of the SARS-CoV-2 XBB Variant Derived from Recombination of Two Omicron Subvariants",
"author": "Tamura",
"doi-asserted-by": "crossref",
"first-page": "2800",
"journal-title": "Nat. Commun.",
"key": "ref_63",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1016/S1473-3099(23)00010-5",
"article-title": "ACE2 Binding and Antibody Evasion in Enhanced Transmissibility of XBB.1.5",
"author": "Yue",
"doi-asserted-by": "crossref",
"first-page": "278",
"journal-title": "Lancet Infect. Dis.",
"key": "ref_64",
"volume": "23",
"year": "2023"
},
{
"key": "ref_65",
"unstructured": "AstraZeneca (2023, August 22). A Phase I/III Randomized, Double Blind Study to Evaluate the Safety, Efficacy and Neutralizing Activity of AZD5156/AZD3152 for Pre Exposure Prophylaxis of COVID 19 in Participants With Conditions Causing Immune Impairment. Sub-Study: Phase II Open Label Sub-Study to Evaluate the Safety, PK, and Neutralizing Activity of AZD3152 for Pre-Exposure Prophylaxis of COVID-19, Available online: https://clinicaltrials.gov/."
},
{
"key": "ref_66",
"unstructured": "(2023, August 22). ECCMID Poster: P2636 The SARS-CoV-2 Monoclonal Antibody AZD3152 Potently Neutralises Historical and Currently Circulating Variants. Available online: https://congresspublication.com/media/oinf1sme/23-04-05_azd3152-preclinical-eccmid-poster_d3-01.pdf?utm_campaign=ECCMID-33rd-European-Congress-of-Clinical-Microbiology-&-Infectious-Diseases."
}
],
"reference-count": 66,
"references-count": 66,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/2076-2607/11/10/2417"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Virology",
"Microbiology (medical)",
"Microbiology"
],
"subtitle": [],
"title": "Prevalence of SARS-CoV-2 Omicron Sublineages and Spike Protein Mutations Conferring Resistance against Monoclonal Antibodies in a Swedish Cohort during 2022–2023",
"type": "journal-article",
"volume": "11"
}