Variant-specific humoral immune response to SARS-CoV-2 escape mutants arising in clinically severe, prolonged infection
et al., medRxiv, doi:10.1101/2024.01.06.24300890, Jan 2024
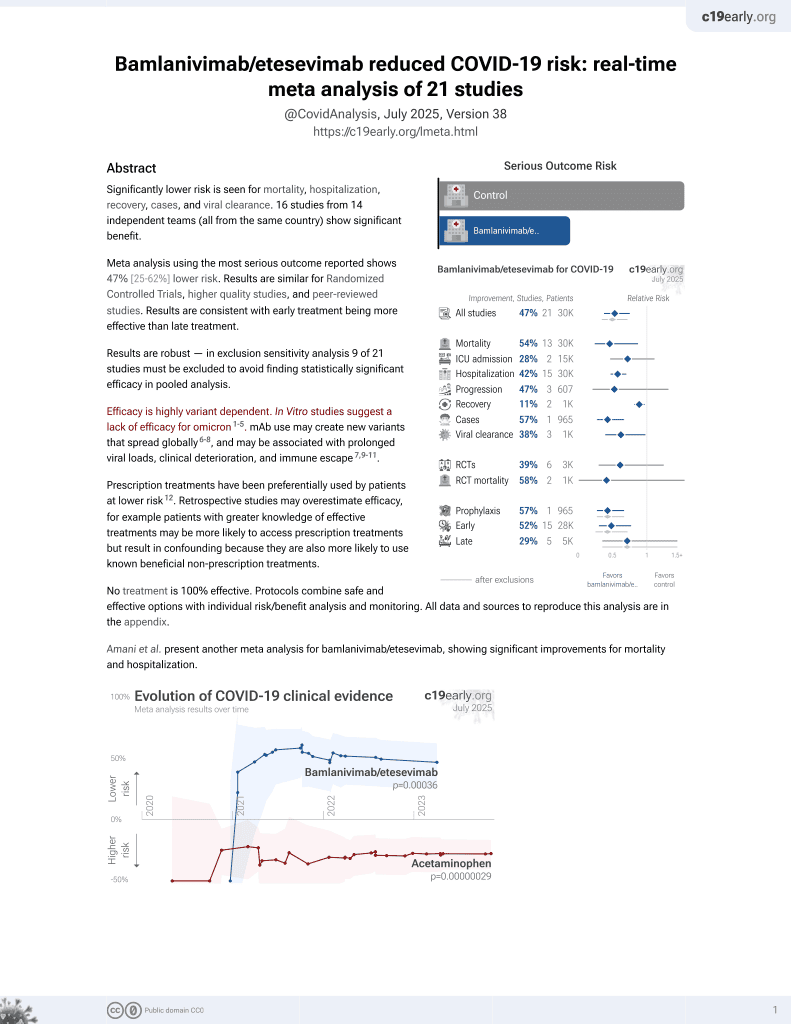
25th treatment shown to reduce risk in
May 2021, now with p = 0.00049 from 22 studies, recognized in 11 countries.
Efficacy is variant dependent.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
Report on a prolonged SARS-CoV-2 infection in an immunocompromised patient treated with bamlanivimab, where bamlanivimab escape variant S:S494P emerged. Later, the onset of an endogenous IgM antibody response coincided with the appearance of additional escape variants carrying mutations in the NTD (S:Delta141-4) and RBD (S:E484K) of the spike protein. The triple mutant S:Delta141-4 E484K S494P became dominant until virus elimination. The study highlights the risk that immunocompromised patients treated with monoclonal antibodies like bamlanivimab can develop escape variants, especially if endogenous antibody responses are insufficient.
1.
Liu et al., Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2, bioRxiv, doi:10.1101/2021.12.14.472719.
2.
Sheward et al., Variable loss of antibody potency against SARS-CoV-2 B.1.1.529 (Omicron), bioRxiv, doi:10.1101/2021.12.19.473354.
3.
VanBlargan et al., An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by several therapeutic monoclonal antibodies, bioRxiv, doi:10.1101/2021.12.15.472828.
Günther et al., 7 Jan 2024, Germany, preprint, 15 authors.
Contact: kuehnj@uni-muenster.de.
Variant-specific humoral immune response to SARS-CoV-2 escape mutants arising in clinically severe, prolonged infection
doi:10.1101/2024.01.06.24300890
Neutralising antibodies against the SARS-CoV-2 spike (S) protein are major determinants of protective immunity, though insufficient antibody responses may cause the emergence of escape mutants. We studied the intra-host evolution and the humoral immune response in a B-cell depleted, haemato-oncologic patient experiencing clinically severe, prolonged SARS-CoV-2 infection with a virus of lineage B.1.177.81. Bamlanivimab treatment early in infection was associated with the emergence of the bamlanivimab escape mutation S:S494P. Ten days before virus elimination, additional mutations within the N-terminal domain (NTD) and the receptor binding domain (RBD) of S were observed, of which the triple mutant S:Delta141-4 E484K S494P became dominant. Routine serology revealed no evidence of an antibody response in the patient. A detailed analysis of the variantspecific immune response by pseudotyped virus neutralisation test (pVNT), surrogate VNT (sVNT), and immunoglobulin-capture EIA showed that the onset of an IgM-dominated antibody response coincided with the appearance of escape mutations. The formation of neutralising antibodies against S:Delta141-4 E484K S494P correlated with virus elimination. One year later, the patient experienced clinically mild re-infection with Omicron BA.1.18, which was treated with sotrovimab and resulted in a massive increase of Omicron-reactive antibodies. In conclusion, the onset of an IgM-dominated endogenous immune response in an immunocompromised patient coincided with the appearance of additional mutations in the NTD and RBD of S in a bamlanivimab-resistant virus. Although virus elimination was ultimately achieved, this humoral immune response escaped detection by routine diagnosis and created a situation temporarily favouring the emergence of escape variants with known epidemiological relevance.
decision to publish. The plasmid pCG1-SARS-2-S was kindly provided by Prof. Dr. Stefan Pöhlmann (Infection Biology Unit, German Primate Centre, Göttingen, Germany). We thank Gert Zimmer (Institute of Virology and Immunology, Mittelhäusern, Switzerland) for the VSV pseudo-typed virus. We acknowledge support from the Open Access Publication Fund of the University of Muenster. All authors read and approved the final version of the manuscript and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Contributors
Declaration of interests The authors declare no conflict of interest related to this work. Parts of this study regarding the methodological establishment of the in-house sVNT and EIA were submitted in 2023 by TG to the Medical Faculty of the University of Muenster, Muenster, Germany, as their doctoral thesis entitled "Etablierung serologischer Nachweisverfahren zur Varianten-spezifischen Detektion und Differenzierung von Antikörpern gegen SARS-CoV-2".
Ethics Statement The patient presented in our study has given informed consent to data collection and publication. The ethics committee of the Aerztekammer Westfalen-Lippe, Muenster, Germany, and the
University of Muenster has ethically approved the study Characterisation of the Humoral Immune Response to SARS-CoV-2 in Vaccinees under the file number 2021-039-f-S. All study..
References
Adams, Hermsen, Lubke, Timm, Schaal, Comparison of commercial SARS-CoV-2 surrogate neutralization assays with a full virus endpoint dilution neutralization test in two different cohorts, J Virol Methods, doi:10.1016/j.jviromet.2022.114569
Alenquer, Ferreira, Lousa, Valerio, Medina-Lopes et al., Signatures in SARS-CoV-2 spike protein conferring escape to neutralizing antibodies, PLoS Pathog, doi:10.1371/journal.ppat.1009772
Avanzato, Matson, Seifert, Pryce, Williamson et al., Case Study: Prolonged Infectious SARS-CoV-2 Shedding from an Asymptomatic Immunocompromised Individual with Cancer, Cell, doi:10.1016/j.cell.2020.10.049
Barnes, West, Huey-Tubman, Hoffmann, Sharaf et al., Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies, Cell, doi:10.1016/j.cell.2020.06.025
Berger Rentsch, Zimmer, A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon, PLoS One, doi:10.1371/journal.pone.0025858
Boshier, Pang, Penner, Parker, Alders et al., Evolution of viral variants in remdesivir-treated and untreated SARS-CoV-2-infected pediatrics patients, J Med Virol, doi:10.1002/jmv.27285
Callaway, Hastie, Schendel, Li, Yu et al., Bivalent intra-spike binding provides durability against emergent Omicron lineages: Results from a global consortium, Cell Rep, doi:10.1016/j.celrep.2023.112014
Cao, Wang, Jian, Song, Yisimayi, Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies, Nature, doi:10.1038/s41586-021-04385-3
Cele, Karim, Lustig, San, Hermanus et al., SARS-CoV-2 prolonged infection during advanced HIV disease evolves extensive immune escape, Cell Host Microbe, doi:10.1016/j.chom.2022.01.005
Cerutti, Guo, Zhou, Gorman, Lee et al., Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite, Cell Host Microbe, doi:10.1016/j.chom.2021.03.005
Chaguza, Hahn, Petrone, Zhou, Ferguson et al., Accelerated SARS-CoV-2 intrahost evolution leading to distinct genotypes during chronic infection, Cell Reports Medicine, doi:10.1016/j.xcrm.2023.100943
Dejnirattisai, Zhou, Ginn, Duyvesteyn, Supasa et al., The antigenic anatomy of SARS-CoV-2 receptor binding domain, Cell, doi:10.1016/j.cell.2021.02.032
Focosi, Maggi, Franchini, Mcconnell, Casadevall, Analysis of Immune Escape Variants from Antibody-Based Therapeutics against COVID-19: A Systematic Review, Int J Mol Sci, doi:10.3390/ijms23010029
Galimidi, Klein, Politzer, Bai, Seaman et al., Intraspike crosslinking overcomes antibody evasion by HIV-1, Cell, doi:10.1016/j.cell.2015.01.016
Gonzalez-Reiche, Alshammary, Schaefer, Patel, Polanco et al., Sequential intrahost evolution and onward transmission of SARS-CoV-2 variants, Nat Commun, doi:10.1038/s41467-023-38867-x
Harari, Tahor, Rutsinsky, Meijer, Miller et al., Drivers of adaptive evolution during chronic SARS-CoV-2 infections, Nat Med, doi:10.1038/s41591-022-01882-4
Heyer, Gunther, Robitaille, Lutgehetmann, Addo et al., Remdesivir-induced emergence of SARS-CoV2 variants in patients with prolonged infection, Cell Rep Med, doi:10.1016/j.xcrm.2022.100735
Hodcroft, Zuber, Nadeau, Vaughan, Crawford et al., Spread of a SARS-CoV-2 variant through Europe in the summer of 2020, Nature, doi:10.1038/s41586-021-03677-y
Iketani, Liu, Guo, Liu, Chan et al., Antibody evasion properties of SARS-CoV-2 Omicron sublineages, Nature, doi:10.1038/s41586-022-04594-4
Jensen, Luebke, Feldt, Keitel, Brandenburger et al., Emergence of the E484K mutation in SARS-COV-2-infected immunocompromised patients treated with bamlanivimab in Germany, Lancet Reg Health Eur, doi:10.1016/j.lanepe.2021.100164
Kemp, Collier, Datir, Ferreira, Gayed et al., SARS-CoV-2 evolution during treatment of chronic infection, Nature, doi:10.1038/s41586-021-03291-y
Khoury, Cromer, Reynaldi, Schlub, Wheatley et al., Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection, Nature Medicine, doi:10.1038/s41591-021-01377-8
King, Bryan, Hilchey, Wang, Zand, First Impressions Matter: Immune Imprinting and Antibody Cross-Reactivity in Influenza and SARS-CoV-2, Pathogens, doi:10.3390/pathogens12020169
Krammer, A correlate of protection for SARS-CoV-2 vaccines is urgently needed, Nature Medicine, doi:10.1038/s41591-021-01432-4
Ku, Xie, Hinton, Liu, Ye et al., Nasal delivery of an IgM offers broad protection from SARS-CoV-2 variants, Nature, doi:10.1038/s41586-021-03673-2
Li, Wu, Nie, Zhang, Hao et al., The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity, Cell, doi:10.1016/j.cell.2020.07.012
Liu, Iketani, Guo, Chan, Wang et al., Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2, Nature, doi:10.1038/s41586-021-04388-0
Lok, An NTD supersite of attack, Cell Host & Microbe, doi:10.1016/j.chom.2021.04.010
Markov, Ghafari, Beer, Lythgoe, Simmonds et al., The evolution of SARS-CoV-2, Nature Reviews Microbiology, doi:10.1038/s41579-023-00878-2
Mccallum, Marco, Lempp, Tortorici, Pinto et al., N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2, Cell, doi:10.1016/j.cell.2021.03.028
Mccarthy, Rennick, Nambulli, Mccarthy, Bain et al., Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape, Science, doi:10.1126/science.abf6950
Mcmahan, Yu, Mercado, Loos, Tostanoski et al., Correlates of protection against SARS-CoV-2 in rhesus macaques, Nature, doi:10.1038/s41586-020-03041-6
Meijers, Ruchnewitz, Eberhardt, Łuksza, Lässig, Population immunity predicts evolutionary trajectories of SARS-CoV-2, Cell, doi:10.1016/j.cell.2023.09.022
Mohamed, Anhlan, Schöfbänker, Schreiber, Classen et al., Hypericum perforatum and Its Ingredients Hypericin and Pseudohypericin Demonstrate an Antiviral Activity against SARS-CoV-2, Pharmaceuticals, doi:10.3390/ph15050530
Muecksch, Weisblum, Barnes, Schmidt, Schaefer-Babajew et al., Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations, Immunity, doi:10.1016/j.immuni.2021.07.008
Piccoli, Park, Tortorici, Czudnochowski, Walls et al., Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology, Cell, doi:10.1016/j.cell.2020.09.037
Qi, Liu, Wang, Zhang, The humoral response and antibodies against SARS-CoV-2 infection, Nat Immunol, doi:10.1038/s41590-022-01248-5
Rambaut, Holmes, 'toole, Hill, Mccrone et al., A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology, Nature Microbiology, doi:10.1038/s41564-020-0770-5
Reynolds, Pade, Gibbons, Otter, Lin et al., Immune boosting by B.1.1.529 (Omicron) depends on previous SARS-CoV-2 exposure, Science, doi:10.1126/science.abq1841
Rockett, Basile, Maddocks, Fong, Agius et al., Resistance Mutations in SARS-CoV-2 Delta Variant after Sotrovimab Use, N Engl J Med, doi:10.1056/NEJMc2120219
Roemer, Sheward, Hisner, Gueli, Sakaguchi et al., SARS-CoV-2 evolution in the Omicron era, Nature Microbiology, doi:10.1038/s41564-023-01504-w
Scheiblauer, Nubling, Wolf, Khodamoradi, Bellinghausen et al., Antibody response to SARS-CoV-2 for more than one year -kinetics and persistence of detection are predominantly determined by avidity progression and test design, J Clin Virol, doi:10.1016/j.jcv.2021.105052
Schoefbaenker, Neddermeyer, Guenther, Mueller, Romberg et al., Surrogate Virus Neutralisation Test Based on Nanoluciferase-Tagged Antigens to Quantify Inhibitory Antibodies against SARS-CoV-2 and Characterise Omicron-Specific Reactivity in a Vaccination Cohort, Vaccines, doi:10.3390/vaccines11121832
Sonnleitner, Prelog, Sonnleitner, Hinterbichler, Halbfurter et al., Cumulative SARS-CoV-2 mutations and corresponding changes in immunity in an immunocompromised patient indicate viral evolution within the host, Nature Communications, doi:10.1038/s41467-022-30163-4
Starr, Greaney, Addetia, Hannon, Choudhary et al., Prospective mapping of viral mutations that escape antibodies used to treat COVID-19, Science, doi:10.1126/science.abf9302
Starr, Greaney, Dingens, Bloom, Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016, Cell Rep Med, doi:10.1016/j.xcrm.2021.100255
Suryadevara, Shrihari, Gilchuk, Vanblargan, Binshtein et al., Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein, Cell, doi:10.1016/j.cell.2021.03.029
Tan, Chia, Qin, Liu, Chen et al., A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction, Nat Biotechnol, doi:10.1038/s41587-020-0631-z
Taylor, Adams, Hufford, De, Torre et al., Neutralizing monoclonal antibodies for treatment of COVID-19, Nature Reviews Immunology, doi:10.1038/s41577-021-00542-x
Taylor, Hurst, Charlton, Bailey, Kanji et al., A New SARS-CoV-2 Dual-Purpose Serology Test: Highly Accurate Infection Tracing and Neutralizing Antibody Response Detection, J Clin Microbiol, doi:10.1128/JCM.02438-20
Team, Past SARS-CoV-2 infection protection against re-infection: a systematic review and meta-analysis, Lancet, doi:10.1016/S0140-6736(22)02465-5
Truong, Ryutov, Pandey, Yee, Goldberg et al., Increased viral variants in children and young adults with impaired humoral immunity and persistent SARS-CoV-2 infection: A consecutive case series, EBioMedicine, doi:10.1016/j.ebiom.2021.103355
Voloch, Da, Francisco, De Almeida, Brustolini et al., Intra-host evolution during SARS-CoV-2 prolonged infection, Virus Evol, doi:10.1093/ve/veab078
Von Rhein, Scholz, Henss, Kronstein-Wiedemann, Schwarz et al., Comparison of potency assays to assess SARS-CoV-2 neutralizing antibody capacity in COVID-19 convalescent plasma, J Virol Methods, doi:10.1016/j.jviromet.2020.114031
Wang, Nair, Liu, Iketani, Luo et al., Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7, Nature, doi:10.1038/s41586-021-03398-2
Weigang, Fuchs, Zimmer, Schnepf, Kern et al., Within-host evolution of SARS-CoV-2 in an immunosuppressed COVID-19 patient as a source of immune escape variants, Nature Communications, doi:10.1038/s41467-021-26602-3
Widera, Wilhelm, Hoehl, Pallas, Kohmer et al., Limited Neutralization of Authentic Severe Acute Respiratory Syndrome Coronavirus 2 Variants Carrying E484K In Vitro, J Infect Dis, doi:10.1093/infdis/jiab355
Wise, Covid-19: Omicron infection is poor booster to immunity, study finds, BMJ, doi:10.1136/bmj.o1474
Zahradník, Marciano, Shemesh, Zoler, Harari et al., SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution, Nature Microbiology, doi:10.1038/s41564-021-00954-4
DOI record:
{
"DOI": "10.1101/2024.01.06.24300890",
"URL": "http://dx.doi.org/10.1101/2024.01.06.24300890",
"abstract": "<jats:title>Abstract</jats:title><jats:p>Neutralising antibodies against the SARS-CoV-2 spike (S) protein are major determinants of protective immunity, though insufficient antibody responses may cause the emergence of escape mutants. We studied the intra-host evolution and the humoral immune response in a B-cell depleted, haemato-oncologic patient experiencing clinically severe, prolonged SARS-CoV-2 infection with a virus of lineage B.1.177.81.</jats:p><jats:p>Bamlanivimab treatment early in infection was associated with the emergence of the bamlanivimab escape mutation S:S494P. Ten days before virus elimination, additional mutations within the N-terminal domain (NTD) and the receptor binding domain (RBD) of S were observed, of which the triple mutant S:Delta141-4 E484K S494P became dominant. Routine serology revealed no evidence of an antibody response in the patient. A detailed analysis of the variant-specific immune response by pseudotyped virus neutralisation test (pVNT), surrogate VNT (sVNT), and immunoglobulin-capture EIA showed that the onset of an IgM-dominated antibody response coincided with the appearance of escape mutations. The formation of neutralising antibodies against S:Delta141-4 E484K S494P correlated with virus elimination. One year later, the patient experienced clinically mild re-infection with Omicron BA.1.18, which was treated with sotrovimab and resulted in a massive increase of Omicron-reactive antibodies.</jats:p><jats:p>In conclusion, the onset of an IgM-dominated endogenous immune response in an immunocompromised patient coincided with the appearance of additional mutations in the NTD and RBD of S in a bamlanivimab-resistant virus. Although virus elimination was ultimately achieved, this humoral immune response escaped detection by routine diagnosis and created a situation temporarily favouring the emergence of escape variants with known epidemiological relevance.</jats:p>",
"accepted": {
"date-parts": [
[
2024,
1,
7
]
]
},
"author": [
{
"affiliation": [],
"family": "Günther",
"given": "Theresa",
"sequence": "first"
},
{
"affiliation": [],
"family": "Schöfbänker",
"given": "Michael",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5570-1897",
"affiliation": [],
"authenticated-orcid": false,
"family": "Lorentzen",
"given": "Eva Ulla",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Romberg",
"given": "Marie-Luise",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hennies",
"given": "Marc Tim",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Neddermeyer",
"given": "Rieke",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Müller",
"given": "Marlin Maybrit",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Mellmann",
"given": "Alexander",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Lenz",
"given": "Georg",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Stelljes",
"given": "Matthias",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hrincius",
"given": "Eike Roman",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Vollenberg",
"given": "Richard",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ludwig",
"given": "Stephan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Tepasse",
"given": "Phil-Robin",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-2992-1471",
"affiliation": [],
"authenticated-orcid": false,
"family": "Kühn",
"given": "Joachim Ewald",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
1,
10
]
],
"date-time": "2024-01-10T07:15:20Z",
"timestamp": 1704870920000
},
"deposited": {
"date-parts": [
[
2024,
1,
10
]
],
"date-time": "2024-01-10T07:33:16Z",
"timestamp": 1704871996000
},
"group-title": "Infectious Diseases (except HIV/AIDS)",
"indexed": {
"date-parts": [
[
2024,
1,
11
]
],
"date-time": "2024-01-11T00:25:52Z",
"timestamp": 1704932752092
},
"institution": [
{
"name": "medRxiv"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2024,
1,
7
]
]
},
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2024.01.06.24300890",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2024,
1,
7
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2024,
1,
7
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference": [
{
"DOI": "10.1038/s41564-023-01504-w",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.1"
},
{
"DOI": "10.1016/S0140-6736(22)02465-5",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.2"
},
{
"DOI": "10.1038/s41579-023-00878-2",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.3"
},
{
"DOI": "10.1016/j.chom.2022.01.005",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.4"
},
{
"DOI": "10.1016/j.xcrm.2023.100943",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.5"
},
{
"DOI": "10.1038/s41586-020-03041-6",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.6"
},
{
"DOI": "10.1038/s41591-021-01432-4",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.7"
},
{
"DOI": "10.1038/s41591-021-01377-8",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.8"
},
{
"DOI": "10.1016/j.cell.2023.09.022",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.9"
},
{
"DOI": "10.3390/pathogens12020169",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.10"
},
{
"DOI": "10.1126/science.abq1841",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.11"
},
{
"DOI": "10.1136/bmj.o1474",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.12"
},
{
"DOI": "10.1016/j.jviromet.2020.114031",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.13"
},
{
"DOI": "10.1128/JCM.02438-20",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.14"
},
{
"DOI": "10.1038/s41587-020-0631-z",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.15"
},
{
"DOI": "10.3390/vaccines11121832",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.16"
},
{
"DOI": "10.3390/ph15050530",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.17"
},
{
"DOI": "10.1371/journal.pone.0025858",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.18"
},
{
"DOI": "10.1038/s41564-020-0770-5",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.19"
},
{
"DOI": "10.1038/s41586-021-03677-y",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.20"
},
{
"DOI": "10.1016/j.jviromet.2022.114569",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.21"
},
{
"DOI": "10.3390/ijms23010029",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.22"
},
{
"DOI": "10.1056/NEJMc2120219",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.23"
},
{
"DOI": "10.1038/s41586-021-03291-y",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.24"
},
{
"DOI": "10.1016/j.cell.2020.07.012",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.25"
},
{
"DOI": "10.1093/ve/veab078",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.26"
},
{
"DOI": "10.1038/s41467-021-26602-3",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.27"
},
{
"DOI": "10.1038/s41564-021-00954-4",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.28"
},
{
"DOI": "10.1371/journal.ppat.1009772",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.29"
},
{
"DOI": "10.1038/s41577-021-00542-x",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.30"
},
{
"DOI": "10.1038/s41586-021-03398-2",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.31"
},
{
"DOI": "10.1016/j.lanepe.2021.100164",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.32"
},
{
"DOI": "10.1016/j.xcrm.2021.100255",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.33"
},
{
"DOI": "10.1126/science.abf9302",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.34"
},
{
"DOI": "10.1093/infdis/jiab355",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.35"
},
{
"DOI": "10.1038/s41591-022-01882-4",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.36"
},
{
"DOI": "10.1016/j.cell.2020.10.049",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.37"
},
{
"DOI": "10.1038/s41467-023-38867-x",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.38"
},
{
"DOI": "10.1038/s41586-021-04385-3",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.39"
},
{
"DOI": "10.1016/j.chom.2021.04.010",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.40"
},
{
"DOI": "10.1016/j.cell.2021.03.028",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.41"
},
{
"DOI": "10.1016/j.chom.2021.03.005",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.42"
},
{
"DOI": "10.1126/science.abf6950",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.43"
},
{
"DOI": "10.1002/jmv.27285",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.44"
},
{
"DOI": "10.1016/j.xcrm.2022.100735",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.45"
},
{
"DOI": "10.1016/j.jcv.2021.105052",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.46"
},
{
"DOI": "10.1016/j.cell.2020.09.037",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.47"
},
{
"DOI": "10.1038/s41590-022-01248-5",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.48"
},
{
"DOI": "10.1038/s41467-022-30163-4",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.49"
},
{
"DOI": "10.1016/j.ebiom.2021.103355",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.50"
},
{
"DOI": "10.1038/s41586-021-03673-2",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.51"
},
{
"DOI": "10.1016/j.cell.2020.06.025",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.52"
},
{
"DOI": "10.1016/j.celrep.2023.112014",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.53"
},
{
"DOI": "10.1016/j.cell.2015.01.016",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.54"
},
{
"DOI": "10.1016/j.cell.2021.03.029",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.55"
},
{
"DOI": "10.1016/j.cell.2021.02.032",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.56"
},
{
"DOI": "10.1038/s41586-022-04594-4",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.57"
},
{
"DOI": "10.1038/s41586-021-04388-0",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.58"
},
{
"DOI": "10.1016/j.immuni.2021.07.008",
"doi-asserted-by": "publisher",
"key": "2024010922061368000_2024.01.06.24300890v1.59"
}
],
"reference-count": 59,
"references-count": 59,
"relation": {},
"resource": {
"primary": {
"URL": "http://medrxiv.org/lookup/doi/10.1101/2024.01.06.24300890"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subtitle": [],
"subtype": "preprint",
"title": "Variant-specific humoral immune response to SARS-CoV-2 escape mutants arising in clinically severe, prolonged infection",
"type": "posted-content"
}