Molecular and Structural Aspects of Clinically Relevant Mutations of SARS-CoV-2 RNA-Dependent RNA Polymerase in Remdesivir-Treated Patients
et al., Pharmaceuticals, doi:10.3390/ph16081143, Aug 2023
In silico study showing that specific mutations in the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) may reduce the effectiveness of remdesivir treatment in COVID-19 patients. Authors found that patients with the P323L+G671S+M899I or P323L+G671S+L838I+D738Y+K91E RdRp mutations showed a delayed or no response to remdesivir, as confirmed by increased viral load. Molecular docking and thermodynamic analysis revealed that these mutations altered the binding mode and affinity of remdesivir to RdRp. Principal component analysis showed that the mutations reduced the global motions of the Nsp8 domain of RdRp compared to wild-type, which may impact polymerase activity. Dynamic pharmacophore analysis also showed reduced interactions between remdesivir and RNA in the mutant RdRp complexes.
Gratteri et al., 12 Aug 2023, peer-reviewed, 17 authors.
Contact: federica.moraca@unina.it (corresponding author), carmen.gratteri@studenti.unicz.it, gcosta@unicz.it, alcaro@unicz.it, ambrosio@unicz.it, antonio.lupia@unica.it, bruno.catalanotti@unina.it, maria.bellocchi@gmail.com, luca.carioti@yahoo.com, rsalpini@gmail.com, ceccherini@med.uniroma2.it, simone.la.frazia@uniroma2.it, valentina.svicher@uniroma2.it, vincenzo.malagnino@uniroma2.it, loredana.sarmati@uniroma2.it, bryant@inteligand.com, artese@unicz.it.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Molecular and Structural Aspects of Clinically Relevant Mutations of SARS-CoV-2 RNA-Dependent RNA Polymerase in Remdesivir-Treated Patients
Pharmaceuticals, doi:10.3390/ph16081143
1) Background: SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) is a promising therapeutic target to fight COVID-19, and many RdRp inhibitors nucleotide/nucleoside analogs, such as remdesivir, have been identified or are in clinical studies. However, the appearance of resistant mutations could reduce their efficacy. In the present work, we structurally evaluated the impact of RdRp mutations found at baseline in 39 patients treated with remdesivir and associated with a different degree of antiviral response in vivo. (2) Methods: A refined bioinformatics approach was applied to assign SARS-CoV-2 clade and lineage, and to define RdRp mutational profiles. In line with such a method, the same mutations were built and analyzed by combining docking and thermodynamics evaluations with both molecular dynamics and representative pharmacophore models. (3) Results: Clinical studies revealed that patients bearing the most prevalent triple mutant P323L+671S+M899I, which was present in 41% of patients, or the more complex mutational profile P323L+G671S+L838I+D738Y+K91E, which was found with a prevalence of 2.6%, showed a delayed reduced response to remdesivir, as confirmed by the increase in SARS-CoV-2 viral load and by a reduced theoretical binding affinity versus RdRp (∆Gbind WT = -122.70 kcal/mol; ∆Gbind P323L+671S+M899I = -84.78 kcal/mol; ∆Gbind P323L+G671S+L838I+D738Y+K91E = -96.74 kcal/mol). Combined computational approaches helped to rationalize such clinical observations, offering a mechanistic understanding of the allosteric effects of mutants on the global motions of the viral RNA synthesis machine and in the changes of the interactions patterns of remdesivir during its binding.
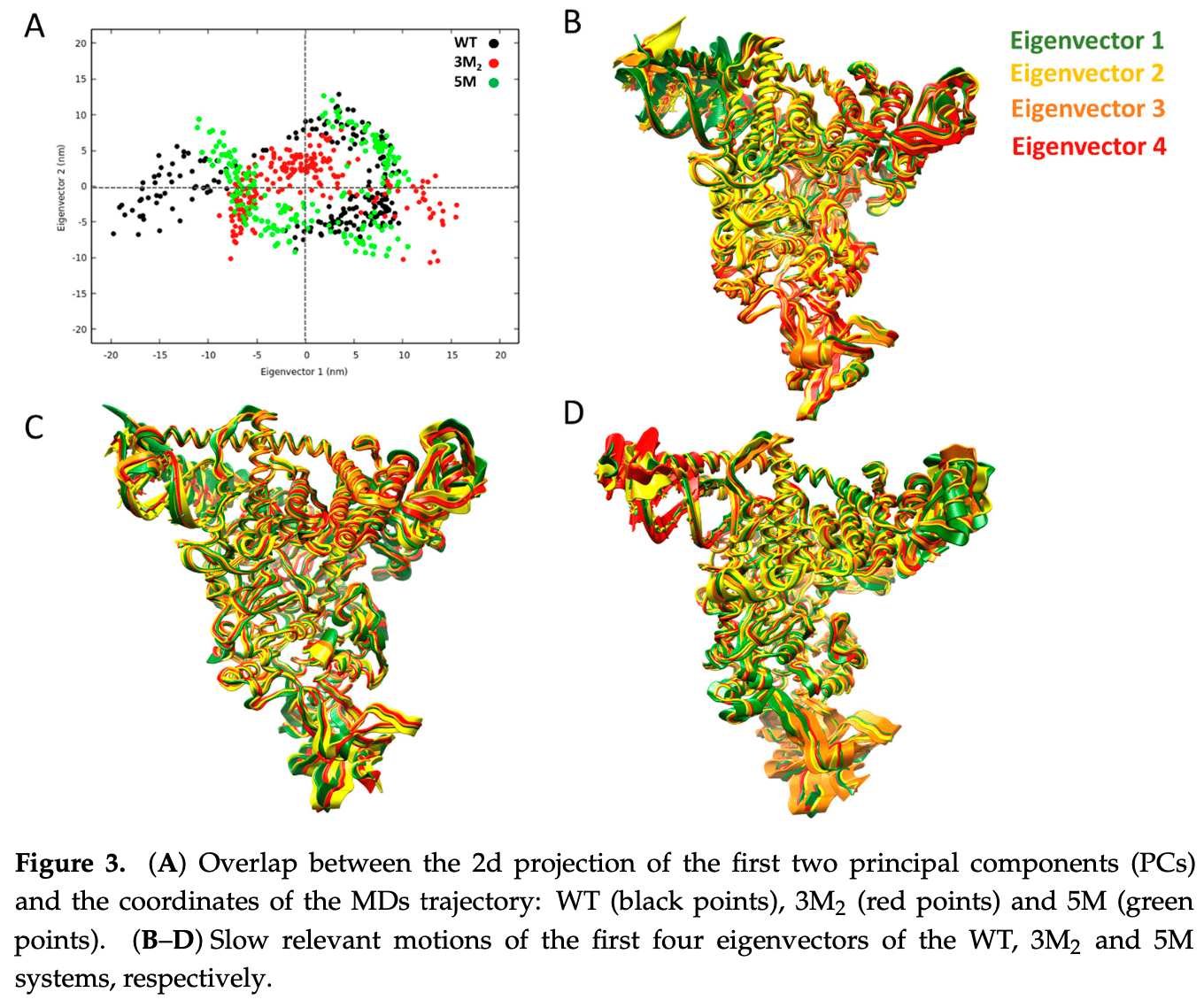
Principal Component Analysis (PCA) Functionally relevant slow motions were analyzed by means of PCA. Specifically, diagonalization of the covariance matrix was performed using the gmx covar tool of GROMACS on the backbone atoms of both the Nsp12/Nsp8/Nsp7/RdRp/NiRAN and the RNA of the WT, 3M 2 , and 5M systems to compare the effect of the mutants with respect to the WT. Then, the motion of the protein was identified by projecting the first four eigenvectors with the gmx anaeig module. Graphs were plotted using the Gnuplot tool.
Pharmacophore Model Generation along the MDs The MDs trajectories, strided every 1 ns, of the three mutated systems (WT, 3M 2 , and 5M), complexed to remdesivir, were used to create representative pharmacophore models (RPMs). RPMs were extracted to understand the dynamic behavior and the chemical features of remdesivir in all of the systems (200 frames for each simulation). This is an innovative technique to reveal the interaction pattern of a ligand in the binding pocket. The advanced version of LigandScout 4.4.9 software [33] was used for the extended pharmacophore investigation on the trajectories of the MDs.
Supplementary Materials: The following supporting information can be downloaded at: https: //www.mdpi.com/article/10.3390/ph16081143/s1 : Figure S1 : flowchart schematically representing the overall approach adopted in the study. Figure S2 : RMSF plot built on the four PCs revealing the changes in the fluctuations of 3M 2 and 5M..
References
Agostini, Andres, Sims, Graham, Sheahan et al., Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease, mBio, doi:10.1128/mBio.00221-18
Agostini, Pruijssers, Chappell, Gribble, Lu et al., Small-molecule antiviral b-d-N4-hydroxycytidine Inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance, J. Virol, doi:10.1128/JVI.01348-19
Aksamentov, Roemer, Hodcroft, Neher, Nextclade: Clade assignment, mutation calling and quality control for viral genomes, J. Open Source Softw, doi:10.21105/joss.03773
Beigel, Tomashek, Dodd, Mehta, Zingman et al., Remdesivir for the treatment of Covid-19-preliminary report, N. Engl. J. Med, doi:10.1056/NEJMoa2007764
Bolger, Lohse, Usadel, Trimmomatic: A flexible trimmer for Illumina sequence data, Bioinformatics, doi:10.1093/bioinformatics/btu170
Bravo, Alonso, Soria, Sánchez Palomino, Sanzo Machuca et al., Genetic Study of SARS-CoV-2 Non Structural Protein 12 in COVID-19 Patients Non Responders to Remdesivir, Microbiol. Spectr, doi:10.1128/spectrum.02448-22
Bravo, Dangerfield, Taylor, Johnson, Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication, Mol. Cell, doi:10.1016/j.molcel.2021.01.035
Byléhn, Menéndez, Perez-Lemus, Alvarado, De Pablo, Modeling the Binding Mechanism of Remdesivir, Favilavir, and Ribavirin to SARS-CoV-2 RNA-Dependent RNA Polymerase, ACS Cent. Sci, doi:10.1021/acscentsci.0c01242
Chen, Liu, Guo, Emerging coronaviruses: Genome structure, replication, and pathogenesis, J. Med. Virol, doi:10.1002/jmv.25681
Cornell, Cieplak, Bayly, Gould, Merz et al., A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules, J. Am. Chem. Soc, doi:10.1021/ja00124a002
Crotty, Maag, Arnold, Zhong, Lau et al., The broadspectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen, Nat. Med, doi:10.1038/82191
Das, Gerlits, Parks, Langan, Kovalevsky et al., Protein kinase A catalytic subunit primed for action: Time-lapse crystallography of michaelis complex formation, Structure, doi:10.1016/j.str.2015.10.005
Friesner, Banks, Murphy, Halgren, Klicic et al., Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy, J. Med. Chem, doi:10.1021/jm0306430
Frisch, Trucks, Schlegel, Scuseria, Robb et al., Revision C.01
Gao, Yan, Huang, Liu, Zhao et al., Structure of the RNA dependent RNA polymerase from COVID-19 virus, Science, doi:10.1126/science.abb7498
Helmy, Fawzy, Elaswad, Sobieh, Kenney et al., The COVID-19 pandemic: A comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control, J. Clin. Med, doi:10.3390/jcm9041225
Hess, Bekker, Berendsen, Fraaije, LINCS: A linear constraint solver for molecular simulations, J. Comput. Chem
Hornak, Abel, Okur, Strockbine, Roitberg et al., Comparison of multiple Amber force fields and development of improved protein backbone parameters, Proteins, doi:10.1002/prot.21123
Ilmjärv, Abdul, Acosta-Gutiérrez, Estarellas, Galdadas et al., Concurrent mutations in RNA-dependent RNA polymerase and spike protein emerged as the epidemiologically most successful SARS-CoV-2 variant, Sci. Rep, doi:10.1038/s41598-021-91662-w
Imbert, Snijder, Dimitrova, Guillemot, Lécine et al., The SARS-Coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein, Virus Res, doi:10.1016/j.virusres.2007.11.017
Jacobson, Pincus, Rapp, Day, Honig et al., A Hierarchical Approach to All-Atom Protein Loop Prediction, Proteins, doi:10.1002/prot.10613
Jorgensen, Maxwell, Tirado-Rives, Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids, J. Am. Chem. Soc, doi:10.1021/ja9621760
Julander, Demarest, Taylor, Gowen, Walling et al., An update on the progress of galidesivir (BCX4430), a broad-spectrum antiviral, Antivir. Res, doi:10.1016/j.antiviral.2021.105180
Kirchdoerfer, Ward, Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors, Nat. Commun, doi:10.1038/s41467-019-10280-3
Lindahl, Abraham, Hess, Van Der Spoel, Gromacs, Source Code; Zenodo
Lupia, Moraca, Bagetta, Maruca, Ambrosio et al., Computerbased techniques for lead identification and optimization II: Advanced search methods, Phys. Sci. Rev, doi:10.1515/psr-2018-0114
Malone, Urakova, Snijder, Campbell, Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design, Nat. Rev. Mol. Cell Biol, doi:10.1038/s41580-021-00432-z
Maruca, Ambrosio, Lupia, Romeo, Rocca et al., Computer-based techniques for lead identification and optimization I: Basics, Phys. Sci. Rev, doi:10.1515/psr-2018-0113
Mcdonald, RNA synthetic mechanisms employed by diverse families of RNA viruses, Wiley Interdiscip. Rev. RNA, doi:10.1002/wrna.1164
Minskaia, Hertzig, Gorbalenya, Campanacci, Cambillau et al., Discovery of an RNA virus 3'->5' exoribonuclease that is critically involved in coronavirus RNA synthesis, Proc. Natl. Acad. Sci, doi:10.1073/pnas.0508200103
Moraca, Marzano, D'amico, Lupia, Di Fonzo et al., Ligand-based drug repurposing strategy identified SARS-CoV-2 RNA G-quadruplex binders, Chem. Commun, doi:10.1039/D2CC03135C
Quasitools, None
Rambaut, Holmes, O'toole, Hill, Mccrone et al., A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology, Nat. Microbiol, doi:10.1038/s41564-020-0770-5
Rehman, Rehman, Yoo, COVID-19 challenges and its therapeutics, Biomed. Pharmacother, doi:10.1016/j.biopha.2021.112015
Salpini, Alkhatib, Costa, Piermatteo, Ambrosio et al., Key genetic elements, single and in clusters, underlying geographically dependent SARS-CoV-2 genetic adaptation and their impact on binding affinity for drugs and immune control, J. Antimicrob. Chemother, doi:10.1093/jac/dkaa444
Sastry, Adzhigirey, Day, Annabhimoju, Sherman, Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments, J. Comput. Aid. Mol. Des, doi:10.1007/s10822-013-9644-8
Schubert, Karousis, Jomaa, Scaiola, Echeverria et al., SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation, Nat. Struct. Mol. Biol, doi:10.1038/s41594-020-0511-8
Shannon, Fattorini, Sama, Selisko, Feracci et al., A dual mechanism of action of AT-527 against Sars-CoV-2 Polymerase, Nat. Commun, doi:10.1038/s41467-022-28113-1
Slanina, Madhugiri, Bylapudi, Schultheiß, Karl et al., Coronavirus replication-transcription complex: Vital and selective NMPylation of a conserved site in nsp9 by the NiRAN-RdRp subunit, Proc. Natl. Acad. Sci, doi:10.1073/pnas.2022310118
Stevens, Pruijssers, Lee, Gordon, Tchesnokov et al., Mutations in the SARS-CoV-2 RNA-dependent RNA polymerase confer resistance to remdesivir by distinct mechanisms, Sci. Transl. Med, doi:10.1126/scitranslmed.abo0718
Subissi, Posthuma, Collet, Zevenhoven-Dobbe, Gorbalenya et al., One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities, Proc. Natl. Acad. Sci, doi:10.1073/pnas.1323705111
Szemiel, Merits, Orton, Maclean, Pinto et al., In vitro selection of Remdesivir resistance suggests evolutionary predictability of SARS-CoV-2, PLoS Pathog, doi:10.1371/journal.ppat.1009929
Thoms, Buschauer, Ameismeier, Koepke, Denk et al., Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2, Science, doi:10.1126/science.abc8665
Träg, Zahn, Improved GAFF2 parameters for fluorinated alkanes and mixed hydro-and fluorocarbons, J. Mol. Model, doi:10.1007/s00894-018-3911-5
Verbist, Thys, Reumers, Wetzels, Van Der Borght et al., A low-frequency virus variant detection pipeline for Illumina sequencing using adaptive base-calling accuracy filtering, Bioinformatics, doi:10.1093/bioinformatics/btu587
Wang, Cieplak, Kollman, How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules?, J. Comput. Chem, doi:10.1002/1096-987X(200009)21:12<1049::AID-JCC3>3.0.CO;2-F
Wang, Wang, Kollman, Case, Automatic Atom Type and Bond Type Perception in Molecular Mechanical Calculations, J. Mol. Graph. Model, doi:10.1016/j.jmgm.2005.12.005
Wang, Wu, Wang, Gao, Liu et al., Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase, Cell, doi:10.1016/j.cell.2020.05.034
Wermuth, Ganellin, Lindberg, Mitscher, Glossary of Terms Used in Medicinal Chemistry, Pure Appl. Chem, doi:10.1351/pac199870051129
Wolber, Langer, LigandScout: 3-D pharmacophores derived from protein-bound Ligands and their use as virtual screening filters, J. Chem. Inf. Model, doi:10.1021/ci049885e
Yin, Mao, Luan, Shen, Shen et al., Structural basis for inhibition of the RNA-dependent RNA polymerase from Sars-CoV-2 by Remdesivir, Science, doi:10.1126/science.abc1560
Zardecki, Dutta, Goodsell, Lowe, Voigt et al., Educational resources supporting molecular explorations through biology and medicine, Protein Sci, doi:10.1002/pro.4200
Zhang, Zhou, Structural Basis of the Potential Binding Mechanism of Remdesivir to SARS-CoV-2 RNA-Dependent RNA Polymerase, J. Phys. Chem. B, doi:10.1021/acs.jpcb.0c04198
Zhou, Chen, Chen, Advances in COVID-19: The virus, the pathogenesis, and evidence based control and therapeutic strategies, Front. Med, doi:10.1007/s11684-020-0773-x
DOI record:
{
"DOI": "10.3390/ph16081143",
"ISSN": [
"1424-8247"
],
"URL": "http://dx.doi.org/10.3390/ph16081143",
"abstract": "<jats:p>(1) Background: SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) is a promising therapeutic target to fight COVID-19, and many RdRp inhibitors nucleotide/nucleoside analogs, such as remdesivir, have been identified or are in clinical studies. However, the appearance of resistant mutations could reduce their efficacy. In the present work, we structurally evaluated the impact of RdRp mutations found at baseline in 39 patients treated with remdesivir and associated with a different degree of antiviral response in vivo. (2) Methods: A refined bioinformatics approach was applied to assign SARS-CoV-2 clade and lineage, and to define RdRp mutational profiles. In line with such a method, the same mutations were built and analyzed by combining docking and thermodynamics evaluations with both molecular dynamics and representative pharmacophore models. (3) Results: Clinical studies revealed that patients bearing the most prevalent triple mutant P323L+671S+M899I, which was present in 41% of patients, or the more complex mutational profile P323L+G671S+L838I+D738Y+K91E, which was found with a prevalence of 2.6%, showed a delayed reduced response to remdesivir, as confirmed by the increase in SARS-CoV-2 viral load and by a reduced theoretical binding affinity versus RdRp (ΔGbindWT = −122.70 kcal/mol; ΔGbindP323L+671S+M899I = −84.78 kcal/mol; ΔGbindP323L+G671S+L838I+D738Y+K91E = −96.74 kcal/mol). Combined computational approaches helped to rationalize such clinical observations, offering a mechanistic understanding of the allosteric effects of mutants on the global motions of the viral RNA synthesis machine and in the changes of the interactions patterns of remdesivir during its binding.</jats:p>",
"alternative-id": [
"ph16081143"
],
"author": [
{
"ORCID": "http://orcid.org/0009-0007-3527-2080",
"affiliation": [
{
"name": "Dipartimento di Scienze della Salute, Campus “S. Venuta”, Università degli Studi “Magna Græcia” di Catanzaro, Viale Europa, 88100 Catanzaro, Italy"
}
],
"authenticated-orcid": false,
"family": "Gratteri",
"given": "Carmen",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0003-4874-2946",
"affiliation": [
{
"name": "Dipartimento di Medicina Sperimentale e Clinica, Campus “S. Venuta”, Università degli Studi “Magna Græcia” di Catanzaro, Viale Europa, 88100 Catanzaro, Italy"
}
],
"authenticated-orcid": false,
"family": "Ambrosio",
"given": "Francesca Alessandra",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-0870-2376",
"affiliation": [
{
"name": "Dipartimento di Scienze della vita e dell’ambiente, Università degli Studi di Cagliari, Cittadella Universitaria di Monserrato, 09124 Cagliari, Italy"
},
{
"name": "Net4Science S.r.l., Università degli Studi “Magna Græcia” di Catanzaro, 88100 Catanzaro, Italy"
}
],
"authenticated-orcid": false,
"family": "Lupia",
"given": "Antonio",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-1077-1971",
"affiliation": [
{
"name": "Net4Science S.r.l., Università degli Studi “Magna Græcia” di Catanzaro, 88100 Catanzaro, Italy"
},
{
"name": "Dipartimento di Farmacia, Università degli Studi di Napoli “Federico II”, Via D. Montesano 49, 80131 Napoli, Italy"
}
],
"authenticated-orcid": false,
"family": "Moraca",
"given": "Federica",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Dipartimento di Farmacia, Università degli Studi di Napoli “Federico II”, Via D. Montesano 49, 80131 Napoli, Italy"
}
],
"family": "Catalanotti",
"given": "Bruno",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-0947-9479",
"affiliation": [
{
"name": "Dipartimento di Scienze della Salute, Campus “S. Venuta”, Università degli Studi “Magna Græcia” di Catanzaro, Viale Europa, 88100 Catanzaro, Italy"
},
{
"name": "Net4Science S.r.l., Università degli Studi “Magna Græcia” di Catanzaro, 88100 Catanzaro, Italy"
}
],
"authenticated-orcid": false,
"family": "Costa",
"given": "Giosuè",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Dipartimento di Medicina Sperimentale, Università Tor Vergata di Roma, Via Montpellier, 1, 00133 Roma, Italy"
}
],
"family": "Bellocchi",
"given": "Maria",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-1713-8682",
"affiliation": [
{
"name": "Dipartimento di Medicina Sperimentale, Università Tor Vergata di Roma, Via Montpellier, 1, 00133 Roma, Italy"
}
],
"authenticated-orcid": false,
"family": "Carioti",
"given": "Luca",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Dipartimento di Medicina Sperimentale, Università Tor Vergata di Roma, Via Montpellier, 1, 00133 Roma, Italy"
}
],
"family": "Salpini",
"given": "Romina",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Dipartimento di Medicina Sperimentale, Università Tor Vergata di Roma, Via Montpellier, 1, 00133 Roma, Italy"
}
],
"family": "Ceccherini-Silberstein",
"given": "Francesca",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Dipartimento di Biologia, Università Tor Vergata di Roma, Via della Ricerca Scientifica, 1, 00133 Roma, Italy"
}
],
"family": "Frazia",
"given": "Simone La",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Dipartimento di Medicina dei Sistemi, Università Tor Vergata di Roma, Via Montpellier, 1, 00133 Roma, Italy"
}
],
"family": "Malagnino",
"given": "Vincenzo",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Dipartimento di Medicina dei Sistemi, Università Tor Vergata di Roma, Via Montpellier, 1, 00133 Roma, Italy"
}
],
"family": "Sarmati",
"given": "Loredana",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Dipartimento di Biologia, Università Tor Vergata di Roma, Via della Ricerca Scientifica, 1, 00133 Roma, Italy"
}
],
"family": "Svicher",
"given": "Valentina",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Inte:Ligand GmbH, Mariahilferstrasse 74B/11, 1070 Vienna, Austria"
}
],
"family": "Bryant",
"given": "Sharon",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-4638-7760",
"affiliation": [
{
"name": "Dipartimento di Scienze della Salute, Campus “S. Venuta”, Università degli Studi “Magna Græcia” di Catanzaro, Viale Europa, 88100 Catanzaro, Italy"
},
{
"name": "Net4Science S.r.l., Università degli Studi “Magna Græcia” di Catanzaro, 88100 Catanzaro, Italy"
}
],
"authenticated-orcid": false,
"family": "Artese",
"given": "Anna",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-0437-358X",
"affiliation": [
{
"name": "Dipartimento di Scienze della Salute, Campus “S. Venuta”, Università degli Studi “Magna Græcia” di Catanzaro, Viale Europa, 88100 Catanzaro, Italy"
},
{
"name": "Net4Science S.r.l., Università degli Studi “Magna Græcia” di Catanzaro, 88100 Catanzaro, Italy"
}
],
"authenticated-orcid": false,
"family": "Alcaro",
"given": "Stefano",
"sequence": "additional"
}
],
"container-title": "Pharmaceuticals",
"container-title-short": "Pharmaceuticals",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2023,
8,
14
]
],
"date-time": "2023-08-14T14:13:57Z",
"timestamp": 1692022437000
},
"deposited": {
"date-parts": [
[
2023,
8,
14
]
],
"date-time": "2023-08-14T15:38:51Z",
"timestamp": 1692027531000
},
"indexed": {
"date-parts": [
[
2023,
8,
14
]
],
"date-time": "2023-08-14T16:11:14Z",
"timestamp": 1692029474382
},
"is-referenced-by-count": 0,
"issue": "8",
"issued": {
"date-parts": [
[
2023,
8,
12
]
]
},
"journal-issue": {
"issue": "8",
"published-online": {
"date-parts": [
[
2023,
8
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
8,
12
]
],
"date-time": "2023-08-12T00:00:00Z",
"timestamp": 1691798400000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/1424-8247/16/8/1143/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "1143",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2023,
8,
12
]
]
},
"published-online": {
"date-parts": [
[
2023,
8,
12
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.3390/jcm9041225",
"doi-asserted-by": "crossref",
"key": "ref_1",
"unstructured": "Helmy, Y.A., Fawzy, M., Elaswad, A., Sobieh, A., Kenney, S.P., and Shehata, A.A. (2020). The COVID-19 pandemic: A comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J. Clin. Med., 9."
},
{
"DOI": "10.1002/jmv.25681",
"article-title": "Emerging coronaviruses: Genome structure, replication, and pathogenesis",
"author": "Chen",
"doi-asserted-by": "crossref",
"first-page": "418",
"journal-title": "J. Med. Virol.",
"key": "ref_2",
"volume": "92",
"year": "2020"
},
{
"DOI": "10.1007/s11684-020-0773-x",
"article-title": "Advances in COVID-19: The virus, the pathogenesis, and evidence based control and therapeutic strategies",
"author": "Zhou",
"doi-asserted-by": "crossref",
"first-page": "117",
"journal-title": "Front. Med.",
"key": "ref_3",
"volume": "14",
"year": "2020"
},
{
"DOI": "10.1126/science.abc8665",
"article-title": "Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2",
"author": "Thoms",
"doi-asserted-by": "crossref",
"first-page": "1249",
"journal-title": "Science",
"key": "ref_4",
"volume": "369",
"year": "2020"
},
{
"DOI": "10.1038/s41594-020-0511-8",
"article-title": "SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation",
"author": "Schubert",
"doi-asserted-by": "crossref",
"first-page": "959",
"journal-title": "Nat. Struct. Mol. Biol.",
"key": "ref_5",
"volume": "27",
"year": "2020"
},
{
"DOI": "10.1016/j.virusres.2007.11.017",
"article-title": "The SARS-Coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein",
"author": "Imbert",
"doi-asserted-by": "crossref",
"first-page": "136",
"journal-title": "Virus Res.",
"key": "ref_6",
"volume": "133",
"year": "2008"
},
{
"DOI": "10.1002/pro.4200",
"article-title": "Educational resources supporting molecular explorations through biology and medicine",
"author": "Zardecki",
"doi-asserted-by": "crossref",
"first-page": "129",
"journal-title": "Protein Sci.",
"key": "ref_7",
"volume": "31",
"year": "2022"
},
{
"DOI": "10.1126/science.abb7498",
"article-title": "Structure of the RNA dependent RNA polymerase from COVID-19 virus",
"author": "Gao",
"doi-asserted-by": "crossref",
"first-page": "779",
"journal-title": "Science",
"key": "ref_8",
"volume": "368",
"year": "2020"
},
{
"DOI": "10.1038/s41467-019-10280-3",
"article-title": "Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors",
"author": "Kirchdoerfer",
"doi-asserted-by": "crossref",
"first-page": "2342",
"journal-title": "Nat. Commun.",
"key": "ref_9",
"volume": "10",
"year": "2019"
},
{
"DOI": "10.1002/wrna.1164",
"article-title": "RNA synthetic mechanisms employed by diverse families of RNA viruses",
"author": "McDonald",
"doi-asserted-by": "crossref",
"first-page": "351",
"journal-title": "Wiley Interdiscip. Rev. RNA",
"key": "ref_10",
"volume": "4",
"year": "2013"
},
{
"DOI": "10.1073/pnas.1323705111",
"article-title": "One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities",
"author": "Subissi",
"doi-asserted-by": "crossref",
"first-page": "E3900",
"journal-title": "Proc. Natl. Acad. Sci.USA",
"key": "ref_11",
"volume": "111",
"year": "2014"
},
{
"DOI": "10.1016/j.cell.2020.05.034",
"article-title": "Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "417",
"journal-title": "Cell",
"key": "ref_12",
"volume": "182",
"year": "2020"
},
{
"DOI": "10.1016/j.str.2015.10.005",
"article-title": "Protein kinase A catalytic subunit primed for action: Time-lapse crystallography of michaelis complex formation",
"author": "Das",
"doi-asserted-by": "crossref",
"first-page": "2331",
"journal-title": "Structure",
"key": "ref_13",
"volume": "23",
"year": "2015"
},
{
"DOI": "10.1128/JVI.01348-19",
"article-title": "Small-molecule antiviral b-d-N4-hydroxycytidine Inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance",
"author": "Agostini",
"doi-asserted-by": "crossref",
"first-page": "e01348-19",
"journal-title": "J. Virol.",
"key": "ref_14",
"volume": "93",
"year": "2019"
},
{
"DOI": "10.1038/82191",
"article-title": "The broadspectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen",
"author": "Crotty",
"doi-asserted-by": "crossref",
"first-page": "1375",
"journal-title": "Nat. Med.",
"key": "ref_15",
"volume": "6",
"year": "2000"
},
{
"DOI": "10.1073/pnas.0508200103",
"article-title": "Discovery of an RNA virus 3’->5’ exoribonuclease that is critically involved in coronavirus RNA synthesis",
"author": "Minskaia",
"doi-asserted-by": "crossref",
"first-page": "5108",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_16",
"volume": "103",
"year": "2006"
},
{
"DOI": "10.1128/mBio.00221-18",
"article-title": "Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease",
"author": "Agostini",
"doi-asserted-by": "crossref",
"first-page": "e00221-e18",
"journal-title": "mBio",
"key": "ref_17",
"volume": "9",
"year": "2018"
},
{
"DOI": "10.1371/journal.ppat.1009929",
"doi-asserted-by": "crossref",
"key": "ref_18",
"unstructured": "Szemiel, A.M., Merits, A., Orton, R.J., MacLean, O.A., Pinto, R.M., Wickenhagen, A., Lieber, G., Turnbull, M.L., Wang, S., and Furnon, W. (2021). In vitro selection of Remdesivir resistance suggests evolutionary predictability of SARS-CoV-2. PLoS Pathog., 17."
},
{
"DOI": "10.1126/scitranslmed.abo0718",
"article-title": "Mutations in the SARS-CoV-2 RNA-dependent RNA polymerase confer resistance to remdesivir by distinct mechanisms",
"author": "Stevens",
"doi-asserted-by": "crossref",
"first-page": "eabo0718",
"journal-title": "Sci. Transl. Med.",
"key": "ref_19",
"volume": "14",
"year": "2022"
},
{
"DOI": "10.1016/j.molcel.2021.01.035",
"article-title": "Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication",
"author": "Bravo",
"doi-asserted-by": "crossref",
"first-page": "1548",
"journal-title": "Mol. Cell",
"key": "ref_20",
"volume": "81",
"year": "2021"
},
{
"DOI": "10.1056/NEJMoa2007764",
"article-title": "Remdesivir for the treatment of Covid-19-preliminary report",
"author": "Beigel",
"doi-asserted-by": "crossref",
"first-page": "1813",
"journal-title": "N. Engl. J. Med.",
"key": "ref_21",
"volume": "383",
"year": "2020"
},
{
"DOI": "10.1056/NEJMoa2023184",
"doi-asserted-by": "crossref",
"key": "ref_22",
"unstructured": "WHO Solidarity Trial Consortium (2021). Repurposed antiviral drugs for Covid-19-interim WHO solidarity trial results. N. Engl. J. Med., 384, 497–511."
},
{
"DOI": "10.1016/j.biopha.2021.112015",
"article-title": "COVID-19 challenges and its therapeutics",
"author": "Rehman",
"doi-asserted-by": "crossref",
"first-page": "112015",
"journal-title": "Biomed. Pharmacother.",
"key": "ref_23",
"volume": "142",
"year": "2021"
},
{
"DOI": "10.1016/j.antiviral.2021.105180",
"article-title": "An update on the progress of galidesivir (BCX4430), a broad-spectrum antiviral",
"author": "Julander",
"doi-asserted-by": "crossref",
"first-page": "105180",
"journal-title": "Antivir. Res.",
"key": "ref_24",
"volume": "195",
"year": "2021"
},
{
"DOI": "10.1021/acs.jpcb.0c04198",
"article-title": "Structural Basis of the Potential Binding Mechanism of Remdesivir to SARS-CoV-2 RNA-Dependent RNA Polymerase",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "6955",
"journal-title": "J. Phys. Chem. B",
"key": "ref_25",
"volume": "124",
"year": "2020"
},
{
"DOI": "10.1038/s41580-021-00432-z",
"article-title": "Structures and functions of coronavirus replication–transcription complexes and their relevance for SARS-CoV-2 drug design",
"author": "Malone",
"doi-asserted-by": "crossref",
"first-page": "21",
"journal-title": "Nat. Rev. Mol. Cell Biol.",
"key": "ref_26",
"volume": "23",
"year": "2022"
},
{
"DOI": "10.1073/pnas.2022310118",
"article-title": "Coronavirus replication-transcription complex: Vital and selective NMPylation of a conserved site in nsp9 by the NiRAN-RdRp subunit",
"author": "Slanina",
"doi-asserted-by": "crossref",
"first-page": "e2022310118",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_27",
"volume": "118",
"year": "2021"
},
{
"DOI": "10.1021/acscentsci.0c01242",
"article-title": "Modeling the Binding Mechanism of Remdesivir, Favilavir, and Ribavirin to SARS-CoV-2 RNA-Dependent RNA Polymerase",
"author": "Alvarado",
"doi-asserted-by": "crossref",
"first-page": "164",
"journal-title": "ACS Cent. Sci.",
"key": "ref_28",
"volume": "7",
"year": "2021"
},
{
"DOI": "10.1351/pac199870051129",
"article-title": "Glossary of Terms Used in Medicinal Chemistry",
"author": "Wermuth",
"doi-asserted-by": "crossref",
"first-page": "1129",
"journal-title": "Pure Appl. Chem.",
"key": "ref_29",
"volume": "70",
"year": "1998"
},
{
"article-title": "Computer-based techniques for lead identification and optimization I: Basics",
"author": "Maruca",
"first-page": "20180113",
"journal-title": "Phys. Sci. Rev.",
"key": "ref_30",
"volume": "4",
"year": "2019"
},
{
"article-title": "Computer-based techniques for lead identification and optimization II: Advanced search methods",
"author": "Lupia",
"first-page": "20180114",
"journal-title": "Phys. Sci. Rev.",
"key": "ref_31",
"volume": "5",
"year": "2019"
},
{
"DOI": "10.1039/D2CC03135C",
"article-title": "Ligand-based drug repurposing strategy identified SARS-CoV-2 RNA G-quadruplex binders",
"author": "Moraca",
"doi-asserted-by": "crossref",
"first-page": "11913",
"journal-title": "Chem. Commun.",
"key": "ref_32",
"volume": "58",
"year": "2022"
},
{
"DOI": "10.1021/ci049885e",
"article-title": "LigandScout: 3-D pharmacophores derived from protein-bound Ligands and their use as virtual screening filters",
"author": "Wolber",
"doi-asserted-by": "crossref",
"first-page": "160",
"journal-title": "J. Chem. Inf. Model.",
"key": "ref_33",
"volume": "45",
"year": "2005"
},
{
"DOI": "10.1126/science.abc1560",
"article-title": "Structural basis for inhibition of the RNA-dependent RNA polymerase from Sars-CoV-2 by Remdesivir",
"author": "Yin",
"doi-asserted-by": "crossref",
"first-page": "1499",
"journal-title": "Science",
"key": "ref_34",
"volume": "368",
"year": "2020"
},
{
"DOI": "10.1038/s41598-021-91662-w",
"article-title": "Concurrent mutations in RNA-dependent RNA polymerase and spike protein emerged as the epidemiologically most successful SARS-CoV-2 variant",
"author": "Abdul",
"doi-asserted-by": "crossref",
"first-page": "13705",
"journal-title": "Sci. Rep.",
"key": "ref_35",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.1093/jac/dkaa444",
"article-title": "Key genetic elements, single and in clusters, underlying geographically dependent SARS-CoV-2 genetic adaptation and their impact on binding affinity for drugs and immune control",
"author": "Salpini",
"doi-asserted-by": "crossref",
"first-page": "396",
"journal-title": "J. Antimicrob. Chemother.",
"key": "ref_36",
"volume": "76",
"year": "2021"
},
{
"DOI": "10.1128/spectrum.02448-22",
"article-title": "Genetic Study of SARS-CoV-2 Non Structural Protein 12 in COVID-19 Patients Non Responders to Remdesivir",
"author": "Alonso",
"doi-asserted-by": "crossref",
"first-page": "e0244822",
"journal-title": "Microbiol. Spectr.",
"key": "ref_37",
"volume": "10",
"year": "2022"
},
{
"key": "ref_38",
"unstructured": "(2023, May 31). Genome Detective Virus Tool. Available online: https://www.genomedetective.com/app/typingtool/virus/."
},
{
"DOI": "10.1093/bioinformatics/btu170",
"article-title": "Trimmomatic: A flexible trimmer for Illumina sequence data",
"author": "Bolger",
"doi-asserted-by": "crossref",
"first-page": "2114",
"journal-title": "Bioinformatics",
"key": "ref_39",
"volume": "30",
"year": "2014"
},
{
"DOI": "10.1093/bioinformatics/btu587",
"article-title": "VirVarSeq: A low-frequency virus variant detection pipeline for Illumina sequencing using adaptive base-calling accuracy filtering",
"author": "Verbist",
"doi-asserted-by": "crossref",
"first-page": "94",
"journal-title": "Bioinformatics",
"key": "ref_40",
"volume": "31",
"year": "2015"
},
{
"key": "ref_41",
"unstructured": "(2023, May 31). Quasitools. Available online: https://phac-nml.github.io/quasitools/."
},
{
"DOI": "10.21105/joss.03773",
"article-title": "Nextclade: Clade assignment, mutation calling and quality control for viral genomes",
"author": "Aksamentov",
"doi-asserted-by": "crossref",
"first-page": "3773",
"journal-title": "J. Open Source Softw.",
"key": "ref_42",
"volume": "6",
"year": "2021"
},
{
"DOI": "10.1038/s41564-020-0770-5",
"article-title": "A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology",
"author": "Rambaut",
"doi-asserted-by": "crossref",
"first-page": "1403",
"journal-title": "Nat. Microbiol.",
"key": "ref_43",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.1038/s41467-022-28113-1",
"article-title": "A dual mechanism of action of AT-527 against Sars-CoV-2 Polymerase",
"author": "Shannon",
"doi-asserted-by": "crossref",
"first-page": "621",
"journal-title": "Nat. Commun.",
"key": "ref_44",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1007/s10822-013-9644-8",
"article-title": "Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments",
"author": "Sastry",
"doi-asserted-by": "crossref",
"first-page": "221",
"journal-title": "J. Comput. Aid. Mol. Des.",
"key": "ref_45",
"volume": "27",
"year": "2013"
},
{
"DOI": "10.1021/ja9621760",
"article-title": "Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids",
"author": "Jorgensen",
"doi-asserted-by": "crossref",
"first-page": "45",
"journal-title": "J. Am. Chem. Soc.",
"key": "ref_46",
"volume": "118",
"year": "1996"
},
{
"DOI": "10.1002/prot.10613",
"article-title": "A Hierarchical Approach to All-Atom Protein Loop Prediction, Proteins, 2004, 55, 351-67",
"author": "Jacobson",
"doi-asserted-by": "crossref",
"first-page": "351",
"journal-title": "Proteins",
"key": "ref_47",
"volume": "55",
"year": "2004"
},
{
"key": "ref_48",
"unstructured": "(2021). Schrödinger Release 2021-4: Maestro, Schrödinger, LLC."
},
{
"DOI": "10.1021/jm0306430",
"article-title": "Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy",
"author": "Friesner",
"doi-asserted-by": "crossref",
"first-page": "1739",
"journal-title": "J. Med. Chem.",
"key": "ref_49",
"volume": "47",
"year": "2004"
},
{
"key": "ref_50",
"unstructured": "(2021). Schrödinger Release 2021-4: MacroModel, Schrödinger, LLC."
},
{
"key": "ref_51",
"unstructured": "Lindahl, E., Abraham, M., Hess, B., and van der Spoel, D. (2021, March 04). GROMACS 2020.6 Source Code; Zenodo. Available online: https://zenodo.org/record/4576060."
},
{
"DOI": "10.1002/1096-987X(200009)21:12<1049::AID-JCC3>3.0.CO;2-F",
"article-title": "How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules?",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "1049",
"journal-title": "J. Comput. Chem.",
"key": "ref_52",
"volume": "21",
"year": "2000"
},
{
"DOI": "10.1016/j.jmgm.2005.12.005",
"article-title": "Automatic Atom Type and Bond Type Perception in Molecular Mechanical Calculations",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "247",
"journal-title": "J. Mol. Graph. Model.",
"key": "ref_53",
"volume": "25",
"year": "2006"
},
{
"key": "ref_54",
"unstructured": "Frisch, M.J., Trucks, G.W., Schlegel, H.B., Scuseria, G.E., Robb, M.A., Cheeseman, J.R., Scalmani, G., Barone, V., Petersson, G.A., and Nakatsuji, H. (2016). Gaussian, Inc.. Available online: https://gaussian.com/citation/."
},
{
"DOI": "10.1007/s00894-018-3911-5",
"article-title": "Improved GAFF2 parameters for fluorinated alkanes and mixed hydro- and fluorocarbons",
"author": "Zahn",
"doi-asserted-by": "crossref",
"first-page": "39",
"journal-title": "J. Mol. Model.",
"key": "ref_55",
"volume": "25",
"year": "2019"
},
{
"DOI": "10.1002/prot.21123",
"article-title": "Comparison of multiple Amber force fields and development of improved protein backbone parameters",
"author": "Hornak",
"doi-asserted-by": "crossref",
"first-page": "712",
"journal-title": "Proteins",
"key": "ref_56",
"volume": "65",
"year": "2006"
},
{
"DOI": "10.1021/ja00124a002",
"article-title": "A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules",
"author": "Cornell",
"doi-asserted-by": "crossref",
"first-page": "5179",
"journal-title": "J. Am. Chem. Soc.",
"key": "ref_57",
"volume": "117",
"year": "1995"
},
{
"DOI": "10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H",
"article-title": "LINCS: A linear constraint solver for molecular simulations",
"author": "Hess",
"doi-asserted-by": "crossref",
"first-page": "1463",
"journal-title": "J. Comput. Chem.",
"key": "ref_58",
"volume": "18",
"year": "1996"
}
],
"reference-count": 58,
"references-count": 58,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/1424-8247/16/8/1143"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Molecular and Structural Aspects of Clinically Relevant Mutations of SARS-CoV-2 RNA-Dependent RNA Polymerase in Remdesivir-Treated Patients",
"type": "journal-article",
"volume": "16"
}