Altered DNA Methylation Pattern Contributes to Differential Epigenetic Immune Signaling in the Upper Respiratory Airway of Unvaccinated COVID-19 Patients
et al., Cells, doi:10.3390/cells14211673, Oct 2025
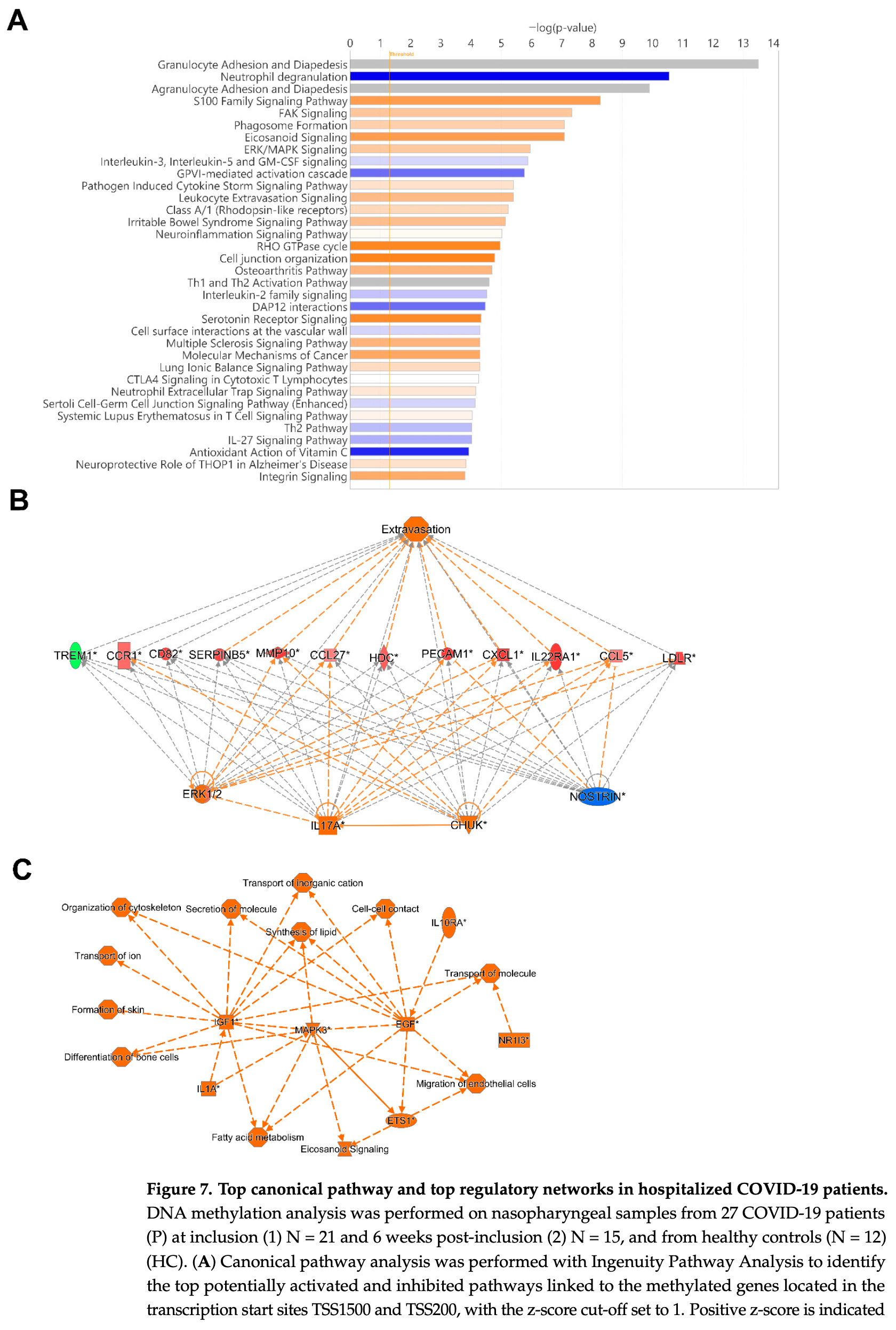
Analysis of DNA methylation patterns in upper respiratory airways of hospitalized COVID-19 patients.
Govender et al., 27 Oct 2025, Sweden, peer-reviewed, 15 authors, study period July 2020 - October 2021.
Contact: melissa.govender@well.ox.ac.uk (corresponding author), francis.hopkins@liu.se, cecilia.svanberg@liu.se, johan.nordgren@liu.se, marie.hagbom@liu.se, jonas.klingstrom@liu.se, sofia.c.nystrom@liu.se, jyotirmoy.das@liu.se, asa.nilsdotter-augustinsson@liu.se, johanna.sjowall@liu.se, ykyong@xmu.edu.my, vvelu@emory.edu, sivraju@gmail.com, shankarem@cutn.ac.in, marie.larsson@liu.se.
Altered DNA Methylation Pattern Contributes to Differential Epigenetic Immune Signaling in the Upper Respiratory Airway of Unvaccinated COVID-19 Patients
Cells, doi:10.3390/cells14211673
What are the main findings? • COVID-19 patients show a unique DNA methylation profile in the upper airway, with over 510,000 differentially methylated CpGs affecting antiviral, interferon, and immune response genes.
• Some methylation changes are temporary, normalizing after 6 weeks, while key immune regulators (e.g., IL17A, ERK1/2, OAS1, MX1) remain significantly involved.
What is the implication of the main finding? • SARS-CoV-2 may reprogram immune and repair pathways in the airways, influencing recovery and susceptibility to future respiratory infections. • These findings provide potential targets for biomarkers and therapeutic strategies to modulate post-COVID-19 airway health.
Author Contributions: Conceptualization, M.L., M.G., V.V., J.N., Å.N.-A., M.H. and J.S.; Methodology, M.G., M.L. and J.D.; Validation, M.G., J.D. and M.L.; Formal Analysis, M.G., J.D., S.N., Y.K.Y., J.K., E.M.S. and M.L.; Investigation, M.G., F.R.H. and C.S.; Resources, Å.N.-A., M.H., J.S., J.N., S.N. and M.L.; Data Curation, M.G., J.D. and M.L.; Writing-Original Draft Preparation, M.G., J.D., E.M.S., S.N., Y.K.Y., V.V., S.R. and M.L.; Writing-Review and Editing, M.G., J.D., E.M.S., J.K., S.N., S.R., V.V., F.R.H., C.S., J.N., Y.K.Y., Å.N.-A., M.H., J.S. and M.L.; Visualization, M.G., J.D. and M.L.; Supervision, M.L. and M.G.; Project Administration, M.L. and M.G.; Funding Acquisition, Å.N.-A., J.S. and M.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement: The studies involving human participants were reviewed and approved by Swedish Ethical Review Authority (Ethics No. 2020-02580). The approval date is 10 June 2020. Informed Consent Statement: All the patients/participants provided written informed consent to participate in this study prior to study enrolment.
Conflicts of Interest: None of the authors have any competing interests in the manuscript.
References
Ackermann, Anders, Bilyy, Bowlin, Daniel et al., Patients with COVID-19: In the dark-NETs of neutrophils, Cell Death Differ, doi:10.1038/s41418-021-00805-z
Al-Qahtani, Pantazi, Alhamlan, Alothaid, Matou-Nasri et al., SARS-CoV-2 modulates inflammatory responses of alveolar epithelial type II cells via PI3K/AKT pathway, Front. Immunol, doi:10.3389/fimmu.2022.1020624
Ayoub, Shaker, Masoud, Hassan, Ezzat et al., Altered expression of serum lncRNA CASC2 and miRNA-21-5p in COVID-19 patients, Hum. Genom, doi:10.1186/s40246-024-00578-9
Balnis, Madrid, Hogan, Drake, Adhikari et al., Whole-Genome Methylation Sequencing Reveals that COVID-19-induced Epigenetic Dysregulation Remains 1 Year after Hospital Discharge, Am. J. Respir. Cell Mol. Biol, doi:10.1165/rcmb.2022-0433LE
Balnis, Madrid, Hogan, Drake, Chieng et al., Blood DNA methylation and COVID-19 outcomes, Clin. Epigenet, doi:10.1186/s13148-021-01102-9
Banerjee, Blanco, Bruce, Honson, Chen et al., SARS-CoV-2 Disrupts Splicing, Translation, and Protein Trafficking to Suppress Host Defenses, Cell, doi:10.1016/j.cell.2020.10.004
Bibert, Guex, Lourenco, Brahier, Papadimitriou-Olivgeris et al., Transcriptomic Signature Differences Between SARS-CoV-2 and Influenza Virus Infected Patients, Front. Immunol, doi:10.3389/fimmu.2021.666163
Biering, De Gomes Sousa, Tjang, Pahmeier, Zhu et al., SARS-CoV-2 Spike triggers barrier dysfunction and vascular leak via integrins and TGF-β signaling, Nat. Commun, doi:10.1038/s41467-022-34910-5
Binkowski, Taryma-Lesniak, Luczkowska, Niedzwiedz, Lechowicz et al., Epigenetic activation of antiviral sensors and effectors of interferon response pathways during SARS-CoV-2 infection, Biomed. Pharmacother, doi:10.1016/j.biopha.2022.113396
Blanco-Melo, Nilsson-Payant, Liu, Uhl, Hoagland et al., Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19, Cell, doi:10.1016/j.cell.2020.04.026
Bradic, Taleb, Thomas, Chidiac, Robay et al., DNA methylation predicts the outcome of COVID-19 patients with acute respiratory distress syndrome, J. Transl. Med, doi:10.1186/s12967-022-03737-5
Carabelli, Peacock, Thorne, Harvey, Hughes et al., SARS-CoV-2 variant biology: Immune escape, transmission and fitness, Nat. Rev. Microbiol, doi:10.1038/s41579-022-00841-7
Cheong, Ravishankar, Sharma, Parkhurst, Grassmann et al., Epigenetic memory of coronavirus infection in innate immune cells and their progenitors, Cell, doi:10.1016/j.cell.2023.07.019
Ciccosanti, Antonioli, Sacchi, Notari, Farina et al., Proteomic analysis identifies a signature of disease severity in the plasma of COVID-19 pneumonia patients associated to neutrophil, platelet and complement activation, Clin. Proteom, doi:10.1186/s12014-022-09377-7
Corley, Pang, Dody, Mudd, Patterson et al., Genome-wide DNA methylation profiling of peripheral blood reveals an epigenetic signature associated with severe COVID-19, J. Leukoc. Biol, doi:10.1002/JLB.5HI0720-466R
De Castro Moura, Davalos, Planas-Serra, Alvarez-Errico, Arribas et al., Epigenome-wide association study of COVID-19 severity with respiratory failure, EBioMedicine, doi:10.1016/j.ebiom.2021.103339
De Morais Batista, Puga, Da Silva, Oliveira, Dos Santos et al., Serum biomarkers associated with SARS-CoV-2 severity, Sci. Rep, doi:10.1038/s41598-022-20062-5
Dey, Vaishak, Deka, Radhakrishnan, Paul et al., Epigenetic perspectives associated with COVID-19 infection and related cytokine storm: An updated review, Infection, doi:10.1007/s15010-023-02017-8
Dhar, Saha, Mitra, Nag Chaudhuri, DNA methylation and regulation of gene expression: Guardian of our health, Nucleus, doi:10.1007/s13237-021-00367-y
Dhingra, Nwanaji-Enwerem, Samet, Ward-Caviness, DNA Methylation Age-Environmental Influences, Health Impacts, and Its Role in Environmental Epidemiology, Curr. Environ. Health Rep, doi:10.1007/s40572-018-0203-2
Ehrlich, DNA hypermethylation in disease: Mechanisms and clinical relevance, Epigenetics, doi:10.1080/15592294.2019.1638701
Espin, Yang, Shannon, Assadian, He et al., Cellular and molecular biomarkers of long COVID: A scoping review, EBioMedicine, doi:10.1016/j.ebiom.2023.104552
Falcao-Holanda, Brunialti, Jasiulionis, Salomao, Epigenetic Regulation in Sepsis, Role in Pathophysiology and Therapeutic Perspective, Front. Med
Fung, Liu, Post-translational modifications of coronavirus proteins: Roles and function, Future Virol, doi:10.2217/fvl-2018-0008
Gallo, Locatello, Mazzoni, Novelli, Annunziato, The central role of the nasal microenvironment in the transmission, modulation, and clinical progression of SARS-CoV-2 infection, Mucosal Immunol, doi:10.1038/s41385-020-00359-2
Giacomelli, Piccarducci, Marchetti, Romei, Martini, Pulmonary fibrosis from molecular mechanisms to therapeutic interventions: Lessons from post-COVID-19 patients, Biochem. Pharmacol, doi:10.1016/j.bcp.2021.114812
Gonzalez-Jaramillo, Portilla-Fernandez, Glisic, Voortman, Ghanbari et al., Epigenetics and Inflammatory Markers: A Systematic Review of the Current Evidence, Int. J. Inflam, doi:10.1155/2019/6273680
Govender, Hopkins, Goransson, Svanberg, Shankar et al., T cell perturbations persist for at least 6 months following hospitalization for COVID-19, Front. Immunol, doi:10.3389/fimmu.2022.931039
Gu, Eils, Schlesner, Complex heatmaps reveal patterns and correlations in multidimensional genomic data, Bioinformatics, doi:10.1093/bioinformatics/btw313
Guimaraes, Rossini, Lameu, Implications of SARS-CoV-2 infection on eNOS and iNOS activity: Consequences for the respiratory and vascular systems, Nitric Oxide, doi:10.1016/j.niox.2021.04.003
Higgins, Nilsson-Payant, Bonaventure, Kurland, Ye et al., SARS-CoV-2 hijacks p38beta/MAPK11 to promote virus replication, mBio, doi:10.1128/mbio.01007-23
Hopkins, Govender, Svanberg, Nordgren, Waller et al., Major alterations to monocyte and dendritic cell subsets lasting more than 6 months after hospitalization for COVID-19, Front. Immunol, doi:10.3389/fimmu.2022.1082912
Hu, Guo, Zhou, Shi, Characteristics of SARS-CoV-2 and COVID-19, Nat. Rev. Microbiol, doi:10.1038/s41579-020-00459-7
Huoman, Sayyab, Apostolou, Karlsson, Porcile et al., Epigenetic rewiring of pathways related to odour perception in immune cells exposed to SARS-CoV-2 in vivo and in vitro, Epigenetics, doi:10.1080/15592294.2022.2089471
Jaffe, Irizarry, Accounting for cellular heterogeneity is critical in epigenome-wide association studies, Genome Biol, doi:10.1186/gb-2014-15-2-r31
Kassambara, Mundt, Factoextra: Extract and Visualize the Results of Multivariate Data Analyses, R Package Version
Kee, Thudium, Renner, Glastad, Palozola et al., SARS-CoV-2 disrupts host epigenetic regulation via histone mimicry, Nature, doi:10.1038/s41586-022-05282-z
Khalil, Elemam, Maghazachi, Chemokines and chemokine receptors during COVID-19 infection, Comput. Struct. Biotechnol. J, doi:10.1016/j.csbj.2021.01.034
Khalil, Epigenetics, None
Konigsberg, Barnes, Campbell, Davidson, Zhen et al., Host methylation predicts SARS-CoV-2 infection and clinical outcome, Commun. Med, doi:10.1038/s43856-021-00042-y
Konwar, Asiimwe, Inkster, Merrill, Negri et al., Risk-focused differences in molecular processes implicated in SARS-CoV-2 infection: Corollaries in DNA methylation and gene expression, Epigenetics Chromatin, doi:10.1186/s13072-021-00428-1
Kudriavtsev, Vakhrusheva, Novossmallie, Bozdaganyan, Shaitan et al., Immune Escape Associated with RBD Omicron Mutations and SARS-CoV-2 Evolution Dynamics, Viruses, doi:10.3390/v14081603
Lu, Zhang, Dauphars, He, A Potential Role of Interleukin 10 in COVID-19 Pathogenesis, Trends Immunol, doi:10.1016/j.it.2020.10.012
Lê, Josse, Husson, Factominer, An R Package for Multivariate Analysis, J. Stat. Softw, doi:10.18637/jss.v025.i01
Mcintosh, COVID-19: Clinical Features UpToDate
Mckennan, Nicolae, Estimating and Accounting for Unobserved Covariates in High-Dimensional Correlated Data, J. Am. Stat. Assoc, doi:10.1080/01621459.2020.1769635
Montazersaheb, Hosseiniyan Khatibi, Hejazi, Tarhriz, Farjami et al., COVID-19 infection: An overview on cytokine storm and related interventions, Virol. J, doi:10.1186/s12985-022-01814-1
Moore, Le, Fan, DNA methylation and its basic function, Neuropsychopharmacology, doi:10.1038/npp.2012.112
Moss, Magenheim, Neiman, Zemmour, Loyfer et al., Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease, Nat. Commun, doi:10.1038/s41467-018-07466-6
Nie, Mou, Long, Deng, Hu et al., SARS-CoV-2 ORF3a positively regulates NF-kappaB activity by enhancing IKKbeta-NEMO interaction, Virus Res, doi:10.1016/j.virusres.2023.199086
Nikesjo, Sayyab, Karlsson, Apostolou, Rosen et al., Defining post-acute COVID-19 syndrome (PACS) by an epigenetic biosignature in peripheral blood mononuclear cells, Clin. Epigenet, doi:10.1186/s13148-022-01398-1
Nordlund, Backlin, Wahlberg, Busche, Berglund et al., Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia, Genome Biol, doi:10.1186/gb-2013-14-9-r105
Paradis, Claude, Strimmer, Ape, Analyses of Phylogenetics and Evolution in R language, Bioinformatics, doi:10.1093/bioinformatics/btg412
Phua, Weng, Ling, Egi, Lim et al., Intensive care management of coronavirus disease 2019 (COVID-19): Challenges and recommendations, Lancet Respir. Med, doi:10.1016/S2213-2600(20)30161-2
Qin, Scicluna, Van Der Poll, The Role of Host Cell DNA Methylation in the Immune Response to Bacterial Infection, Front. Immunol, doi:10.3389/fimmu.2021.696280
Saksena, Reddy, Miranda-Saksena, Cardoso, Silva et al., SARS-CoV-2 variants, its recombinants and epigenomic exploitation of host defenses, Biochim. Et. Biophys. Acta (BBA)-Mol. Basis Dis, doi:10.1016/j.bbadis.2023.166836
Samblas, Milagro, Martinez, DNA methylation markers in obesity, metabolic syndrome, and weight loss, Epigenetics, doi:10.1080/15592294.2019.1595297
Sarker, Rahaman, Islam, Alamin, Husain et al., Identification of host genomic biomarkers from multiple transcriptomics datasets for diagnosis and therapies of SARS-CoV-2 infections, PLoS ONE, doi:10.1371/journal.pone.0281981
Sawalha, Zhao, Coit, Lu, Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients, Clin. Immunol, doi:10.1016/j.clim.2020.108410
Schreiber, Viemann, Schoning, Schloer, Mecate Zambrano et al., The MEK1/2-inhibitor ATR-002 efficiently blocks SARS-CoV-2 propagation and alleviates pro-inflammatory cytokine/chemokine responses, Cell Mol. Life Sci, doi:10.1007/s00018-021-04085-1
Schulte-Schrepping, Reusch, Paclik, Bassler, Schlickeiser et al., Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment, Cell, doi:10.1016/j.cell.2020.08.001
Sharif-Askari, Sharif-Askari, Hafezi, Mdkhana, Alsayed et al., Interleukin-17, a salivary biomarker for COVID-19 severity, PLoS ONE, doi:10.1371/journal.pone.0274841
Silva, Ribeiro, Gouveia, Marcelino, Santos et al., Hyperinflammatory Response in COVID-19: A Systematic Review, Viruses, doi:10.3390/v15020553
Teschendorff, Marabita, Lechner, Bartlett, Tegner et al., A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data, Bioinformatics, doi:10.1093/bioinformatics/bts680
Teschendorff, Zhuang, Widschwendter, Independent surrogate variable analysis to deconvolve confounding factors in large-scale microarray profiling studies, Bioinformatics, doi:10.1093/bioinformatics/btr171
The R Core, Team, R: A Language and Environment for Statistical Computing
Tian, Morris, Webster, Yang, Beck et al., Updated methylation analysis pipeline for Illumina BeadChips, Bioinformatics, doi:10.1093/bioinformatics/btx513
Tirado-Magallanes, Rebbani, Lim, Pradhan, Benoukraf, Whole genome DNA methylation: Beyond genes silencing, Oncotarget, doi:10.18632/oncotarget.13562
Vanderheiden, Ralfs, Chirkova, Upadhyay, Zimmerman et al., Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures, J. Virol, doi:10.1128/JVI.00985-20
Vassiliou, Zacharis, Keskinidou, Jahaj, Pratikaki et al., Soluble Angiotensin Converting Enzyme 2 (ACE2) Is Upregulated and Soluble Endothelial Nitric Oxide Synthase (eNOS) Is Downregulated in COVID-19-induced Acute Respiratory Distress Syndrome (ARDS), Pharmaceuticals, doi:10.3390/ph14070695
Volpe, Das, methylR: A graphical interface for comprehensive DNA methylation array data analysis, Bioinformatics, doi:10.1093/bioinformatics/btad184
Wan, Qiu, Baccarelli, Carey, Bacherman et al., Systemic steroid exposure is associated with differential methylation in chronic obstructive pulmonary disease, Am. J. Respir. Crit. Care Med, doi:10.1164/rccm.201207-1280OC
Weckbach, Schweizer, Kraechan, Bieber, Ishikawa-Ankerhold et al., Association of Complement and MAPK Activation With SARS-CoV-2-Associated Myocardial Inflammation, JAMA Cardiol, doi:10.1001/jamacardio.2021.5133
Who, WHO Coronavirus (COVID-19) Dashboard
Wickham, ggplot2: Elegant Graphics for Data Analysis
Wilk, Rustagi, Zhao, Roque, Martinez-Colon et al., A single-cell atlas of the peripheral immune response in patients with severe COVID-19, Nat. Med, doi:10.1038/s41591-020-0944-y
Winkley, Banerjee, Bradley, Koseva, Cheung et al., Immune cell residency in the nasal mucosa may partially explain respiratory disease severity across the age range, Sci. Rep, doi:10.1038/s41598-021-95532-3
Woodruff, Bonham, Anam, Walker, Faliti et al., Chronic inflammation, neutrophil activity, and autoreactivity splits long COVID, Nat. Commun, doi:10.1038/s41467-023-40012-7
Zhong, Altay, Arif, Edfors, Doganay et al., Next generation plasma proteome profiling of COVID-19 patients with mild to moderate symptoms, eBioMedicine, doi:10.1016/j.ebiom.2021.103723
Zhou, Laird, Shen, Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes, Nucleic Acids Res, doi:10.1093/nar/gkw967
Zhou, Zhang, Dong, Wang, Zhang et al., The role of SARS-CoV-2-mediated NF-kappaB activation in COVID-19 patients, Hypertens. Res, doi:10.1038/s41440-023-01460-2
Zhou, Zhou, Pache, Chang, Khodabakhshi et al., Metascape provides a biologist-oriented resource for the analysis of systems-level datasets, Nat. Commun, doi:10.1038/s41467-019-09234-6
DOI record:
{
"DOI": "10.3390/cells14211673",
"ISSN": [
"2073-4409"
],
"URL": "http://dx.doi.org/10.3390/cells14211673",
"abstract": "<jats:p>SARS-CoV-2 infection remains a global health concern, with its impact on host immune responses not fully understood. In a case–control study, we examined how COVID-19 affects DNA methylation patterns in the upper respiratory airway of hospitalized individuals. DNA methylation arrays were performed on nasopharyngeal samples at inclusion/hospitalization and 6 weeks post-inclusion. We found a distinct DNA methylation pattern in COVID-19 patients compared to healthy controls, identifying 510,099 differentially methylated CpGs. Within the transcription start sites (TSSs) and gene body, COVID-19 patients displayed a higher number of genes/CpGs with elevated methylation levels. Enrichment analysis of TSS-methylated genes revealed effects of SARS-CoV-2 on genes associated with type I interferons, anti-viral and inflammatory responses, and immune functions. Some CpG methylations were transient, and normalized at group level by 6 weeks post-inclusion. Several IFN-regulated genes, including OAS1, OAS3, IFIT3, and MX1, were identified. Among the top regulators were IL17A and ERK1/2, both involved in inflammatory processes. Networks nodes included IGF1 and EGF, associated with processes including tissue repair and activation of immune responses. Overall, our data suggests that COVID-19 can impact the upper airway by modifying gene methylation patterns. This could have implications for conditioning of the airways, how individuals respond to future airway infections, and therapeutic interventions.</jats:p>",
"alternative-id": [
"cells14211673"
],
"author": [
{
"ORCID": "https://orcid.org/0000-0001-8327-5517",
"affiliation": [
{
"name": "Division of Molecular Medicine and Virology, Department of Biomedical and Clinical Sciences, Linköping University, 581 83 Linköping, Sweden"
},
{
"name": "Centre for Human Genetics, Pandemic Sciences Institute, University of Oxford, Oxford OX3 7BN, UK"
}
],
"authenticated-orcid": false,
"family": "Govender",
"given": "Melissa",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0000-0002-5649-4658",
"affiliation": [
{
"name": "Bioinformatics, Core Facility, Division of Cell Biology, Department of Biomedical and Clinical Sciences, Faculty of Medicine and Health Sciences, Linköping University, 581 85 Linköping, Sweden"
},
{
"name": "Clinical Genomics Linköping, SciLife Laboratory, Department of Biomedical and Clinical Sciences, Linköping University, 581 83 Linköping, Sweden"
}
],
"authenticated-orcid": false,
"family": "Das",
"given": "Jyotirmoy",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Division of Molecular Medicine and Virology, Department of Biomedical and Clinical Sciences, Linköping University, 581 83 Linköping, Sweden"
}
],
"family": "Hopkins",
"given": "Francis R.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Division of Molecular Medicine and Virology, Department of Biomedical and Clinical Sciences, Linköping University, 581 83 Linköping, Sweden"
}
],
"family": "Svanberg",
"given": "Cecilia",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-5349-2569",
"affiliation": [
{
"name": "Division of Molecular Medicine and Virology, Department of Biomedical and Clinical Sciences, Linköping University, 581 83 Linköping, Sweden"
}
],
"authenticated-orcid": false,
"family": "Nordgren",
"given": "Johan",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-9770-4623",
"affiliation": [
{
"name": "Division of Molecular Medicine and Virology, Department of Biomedical and Clinical Sciences, Linköping University, 581 83 Linköping, Sweden"
}
],
"authenticated-orcid": false,
"family": "Hagbom",
"given": "Marie",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Division of Molecular Medicine and Virology, Department of Biomedical and Clinical Sciences, Linköping University, 581 83 Linköping, Sweden"
}
],
"family": "Klingström",
"given": "Jonas",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-5719-5601",
"affiliation": [
{
"name": "Division of Infection and Inflammation, Department of Biomedical and Clinical Sciences, Linköping University, 581 83 Linköping, Sweden"
},
{
"name": "Department of Infectious Diseases, Vrinnevi Hospital, 603 79 Norrköping, Sweden"
}
],
"authenticated-orcid": false,
"family": "Nilsdotter-Augustinsson",
"given": "Åsa",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Laboratory Centre, Xiamen University Malaysia, Sepang 43900, Selangor, Malaysia"
}
],
"family": "Yong",
"given": "Yean K.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-4238-1924",
"affiliation": [
{
"name": "Laboratory Medicine, Department of Pathology, School of Medicine, Emory University, Atlanta, GA 30322, USA"
},
{
"name": "Division of Microbiology and Immunology, Emory Vaccine Center, Emory National Primate Research Center, Emory University, Atlanta, GA 30329, USA"
}
],
"authenticated-orcid": false,
"family": "Velu",
"given": "Vijayakumar",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "State Public Health Laboratory, Directorate of Public Health and Preventive Medicine, DMS Campus, Teynampet, Chennai 600 018, Tamil Nadu, India"
}
],
"family": "Raju",
"given": "Sivadoss",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-5622-866X",
"affiliation": [
{
"name": "Division of Infection and Inflammation, Department of Biomedical and Clinical Sciences, Linköping University, 581 83 Linköping, Sweden"
},
{
"name": "Department of Infectious Diseases, Vrinnevi Hospital, 603 79 Norrköping, Sweden"
}
],
"authenticated-orcid": false,
"family": "Sjöwall",
"given": "Johanna",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-7866-9818",
"affiliation": [
{
"name": "Infection and Inflammation, Department of Biotechnology, Central University of Tamil Nadu, Thiruvarur 610 005, Tamil Nadu, India"
}
],
"authenticated-orcid": false,
"family": "Shankar",
"given": "Esaki M.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-0145-4966",
"affiliation": [
{
"name": "Division of Molecular Medicine and Virology, Department of Biomedical and Clinical Sciences, Linköping University, 581 83 Linköping, Sweden"
},
{
"name": "Department of Clinical Immunology and Transfusion Medicine, Linköping University, 583 30 Linköping, Sweden"
}
],
"authenticated-orcid": false,
"family": "Nyström",
"given": "Sofia",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-4524-0177",
"affiliation": [
{
"name": "Division of Molecular Medicine and Virology, Department of Biomedical and Clinical Sciences, Linköping University, 581 83 Linköping, Sweden"
}
],
"authenticated-orcid": false,
"family": "Larsson",
"given": "Marie",
"sequence": "additional"
}
],
"container-title": "Cells",
"container-title-short": "Cells",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
10,
28
]
],
"date-time": "2025-10-28T11:47:19Z",
"timestamp": 1761652039000
},
"deposited": {
"date-parts": [
[
2025,
10,
29
]
],
"date-time": "2025-10-29T05:33:27Z",
"timestamp": 1761716007000
},
"funder": [
{
"DOI": "10.13039/501100004359",
"award": [
"201701091"
],
"award-info": [
{
"award-number": [
"201701091"
]
}
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100004359",
"id-type": "DOI"
}
],
"name": "Swedish Research Council"
},
{
"award": [
"COVID-19 ALF"
],
"award-info": [
{
"award-number": [
"COVID-19 ALF"
]
}
],
"name": "Linköping University Hospital"
},
{
"award": [
"RÖ935411"
],
"award-info": [
{
"award-number": [
"RÖ935411"
]
}
],
"name": "Region Östergötland ALF"
},
{
"name": "Regional ALF Grant 2021"
},
{
"award": [
"ORIP/OD P51OD011132"
],
"award-info": [
{
"award-number": [
"ORIP/OD P51OD011132"
]
}
],
"name": "Emory National Primate Research Center"
}
],
"indexed": {
"date-parts": [
[
2025,
10,
29
]
],
"date-time": "2025-10-29T11:56:51Z",
"timestamp": 1761739011330,
"version": "build-2065373602"
},
"is-referenced-by-count": 0,
"issue": "21",
"issued": {
"date-parts": [
[
2025,
10,
27
]
]
},
"journal-issue": {
"issue": "21",
"published-online": {
"date-parts": [
[
2025,
11
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
10,
27
]
],
"date-time": "2025-10-27T00:00:00Z",
"timestamp": 1761523200000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/2073-4409/14/21/1673/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "1673",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2025,
10,
27
]
]
},
"published-online": {
"date-parts": [
[
2025,
10,
27
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"key": "ref_1",
"unstructured": "CDC (2023, April 11). COVID-19 Treatments and Medications, Available online: https://www.cdc.gov/covid/treatment/?CDC_AAref_Val=https://www.cdc.gov/coronavirus/2019-ncov/your-health/treatments-for-severe-illness.html."
},
{
"key": "ref_2",
"unstructured": "WHO (2023, February 07). WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/."
},
{
"DOI": "10.1097/01.EEM.0000942568.62141.35",
"doi-asserted-by": "crossref",
"key": "ref_3",
"unstructured": "McIntosh, K. (2023, April 11). COVID-19: Clinical Features UpToDate 2023. Available online: https://www.uptodate.com/contents/covid-19-clinical-features."
},
{
"DOI": "10.1016/S2213-2600(20)30161-2",
"article-title": "Intensive care management of coronavirus disease 2019 (COVID-19): Challenges and recommendations",
"author": "Phua",
"doi-asserted-by": "crossref",
"first-page": "506",
"journal-title": "Lancet Respir. Med.",
"key": "ref_4",
"volume": "8",
"year": "2020"
},
{
"article-title": "SARS-CoV-2 variant biology: Immune escape, transmission and fitness",
"author": "Carabelli",
"first-page": "162",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_5",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.20944/preprints202206.0022.v1",
"doi-asserted-by": "crossref",
"key": "ref_6",
"unstructured": "Kudriavtsev, A.V., Vakhrusheva, A.V., Novossmallie, C.V.N., Bozdaganyan, M.E., Shaitan, K.V., Kirpichnikov, M.P., and Sokolova, O.S. (2022). Immune Escape Associated with RBD Omicron Mutations and SARS-CoV-2 Evolution Dynamics. Viruses, 14."
},
{
"DOI": "10.1186/s12985-022-01814-1",
"doi-asserted-by": "crossref",
"key": "ref_7",
"unstructured": "Montazersaheb, S., Hosseiniyan Khatibi, S.M., Hejazi, M.S., Tarhriz, V., Farjami, A., Ghasemian Sorbeni, F., Farahzadi, R., and Ghasemnejad, T. (2022). COVID-19 infection: An overview on cytokine storm and related interventions. Virol. J., 19."
},
{
"DOI": "10.1016/j.ebiom.2021.103723",
"doi-asserted-by": "crossref",
"key": "ref_8",
"unstructured": "Zhong, W., Altay, O., Arif, M., Edfors, F., Doganay, L., Mardinoglu, A., Uhlen, M., and Fagerberg, L. (2021). Next generation plasma proteome profiling of COVID-19 patients with mild to moderate symptoms. eBioMedicine, 74."
},
{
"DOI": "10.1186/s12014-022-09377-7",
"doi-asserted-by": "crossref",
"key": "ref_9",
"unstructured": "Ciccosanti, F., Antonioli, M., Sacchi, A., Notari, S., Farina, A., Beccacece, A., Fusto, M., Vergori, A., D’Offizi, G., and Taglietti, F. (2022). Proteomic analysis identifies a signature of disease severity in the plasma of COVID-19 pneumonia patients associated to neutrophil, platelet and complement activation. Clin. Proteom., 19."
},
{
"DOI": "10.1016/j.biopha.2022.113396",
"doi-asserted-by": "crossref",
"key": "ref_10",
"unstructured": "Binkowski, J., Taryma-Lesniak, O., Luczkowska, K., Niedzwiedz, A., Lechowicz, K., Strapagiel, D., Jarczak, J., Davalos, V., Pujol, A., and Esteller, M. (2022). Epigenetic activation of antiviral sensors and effectors of interferon response pathways during SARS-CoV-2 infection. Biomed. Pharmacother., 153."
},
{
"DOI": "10.1371/journal.pone.0281981",
"doi-asserted-by": "crossref",
"key": "ref_11",
"unstructured": "Sarker, B., Rahaman, M.M., Islam, M.A., Alamin, M.H., Husain, M.M., Ferdousi, F., Ahsan, M.A., and Mollah, M.N.H. (2023). Identification of host genomic biomarkers from multiple transcriptomics datasets for diagnosis and therapies of SARS-CoV-2 infections. PLoS ONE, 18."
},
{
"DOI": "10.3389/fimmu.2021.666163",
"doi-asserted-by": "crossref",
"key": "ref_12",
"unstructured": "Bibert, S., Guex, N., Lourenco, J., Brahier, T., Papadimitriou-Olivgeris, M., Damonti, L., Manuel, O., Liechti, R., Gotz, L., and Tschopp, J. (2021). Transcriptomic Signature Differences Between SARS-CoV-2 and Influenza Virus Infected Patients. Front. Immunol., 12."
},
{
"DOI": "10.3389/fimmu.2022.931039",
"doi-asserted-by": "crossref",
"key": "ref_13",
"unstructured": "Govender, M., Hopkins, F.R., Goransson, R., Svanberg, C., Shankar, E.M., Hjorth, M., Nilsdotter-Augustinsson, A., Sjowall, J., Nystrom, S., and Larsson, M. (2022). T cell perturbations persist for at least 6 months following hospitalization for COVID-19. Front. Immunol., 13."
},
{
"DOI": "10.3389/fimmu.2022.1082912",
"doi-asserted-by": "crossref",
"key": "ref_14",
"unstructured": "Hopkins, F.R., Govender, M., Svanberg, C., Nordgren, J., Waller, H., Nilsdotter-Augustinsson, A., Henningsson, A.J., Hagbom, M., Sjowall, J., and Nystrom, S. (2022). Major alterations to monocyte and dendritic cell subsets lasting more than 6 months after hospitalization for COVID-19. Front. Immunol., 13."
},
{
"DOI": "10.1186/s12967-022-03737-5",
"doi-asserted-by": "crossref",
"key": "ref_15",
"unstructured": "Bradic, M., Taleb, S., Thomas, B., Chidiac, O., Robay, A., Hassan, N., Malek, J., Ait Hssain, A., and Abi Khalil, C. (2022). DNA methylation predicts the outcome of COVID-19 patients with acute respiratory distress syndrome. J. Transl. Med., 20."
},
{
"DOI": "10.1007/s15010-023-02017-8",
"article-title": "Epigenetic perspectives associated with COVID-19 infection and related cytokine storm: An updated review",
"author": "Dey",
"doi-asserted-by": "crossref",
"first-page": "1603",
"journal-title": "Infection",
"key": "ref_16",
"volume": "51",
"year": "2023"
},
{
"DOI": "10.1186/s13148-022-01398-1",
"doi-asserted-by": "crossref",
"key": "ref_17",
"unstructured": "Nikesjo, F., Sayyab, S., Karlsson, L., Apostolou, E., Rosen, A., Hedman, K., and Lerm, M. (2022). Defining post-acute COVID-19 syndrome (PACS) by an epigenetic biosignature in peripheral blood mononuclear cells. Clin. Epigenet., 14."
},
{
"DOI": "10.3389/fimmu.2021.696280",
"doi-asserted-by": "crossref",
"key": "ref_18",
"unstructured": "Qin, W., Scicluna, B.P., and van der Poll, T. (2021). The Role of Host Cell DNA Methylation in the Immune Response to Bacterial Infection. Front. Immunol., 12."
},
{
"DOI": "10.1038/npp.2012.112",
"article-title": "DNA methylation and its basic function",
"author": "Moore",
"doi-asserted-by": "crossref",
"first-page": "23",
"journal-title": "Neuropsychopharmacology",
"key": "ref_19",
"volume": "38",
"year": "2013"
},
{
"DOI": "10.1007/s13237-021-00367-y",
"article-title": "DNA methylation and regulation of gene expression: Guardian of our health",
"author": "Dhar",
"doi-asserted-by": "crossref",
"first-page": "259",
"journal-title": "Nucleus",
"key": "ref_20",
"volume": "64",
"year": "2021"
},
{
"DOI": "10.1007/s40572-018-0203-2",
"article-title": "DNA Methylation Age-Environmental Influences, Health Impacts, and Its Role in Environmental Epidemiology",
"author": "Dhingra",
"doi-asserted-by": "crossref",
"first-page": "317",
"journal-title": "Curr. Environ. Health Rep.",
"key": "ref_21",
"volume": "5",
"year": "2018"
},
{
"DOI": "10.1080/15592294.2019.1595297",
"article-title": "DNA methylation markers in obesity, metabolic syndrome, and weight loss",
"author": "Samblas",
"doi-asserted-by": "crossref",
"first-page": "421",
"journal-title": "Epigenetics",
"key": "ref_22",
"volume": "14",
"year": "2019"
},
{
"key": "ref_23",
"unstructured": "Khalil, C.A. (2021). Medical Epigenetics, Academic Press."
},
{
"DOI": "10.1080/15592294.2022.2089471",
"article-title": "Epigenetic rewiring of pathways related to odour perception in immune cells exposed to SARS-CoV-2 in vivo and in vitro",
"author": "Huoman",
"doi-asserted-by": "crossref",
"first-page": "1875",
"journal-title": "Epigenetics",
"key": "ref_24",
"volume": "17",
"year": "2022"
},
{
"DOI": "10.1038/s43856-021-00042-y",
"doi-asserted-by": "crossref",
"key": "ref_25",
"unstructured": "Konigsberg, I.R., Barnes, B., Campbell, M., Davidson, E., Zhen, Y., Pallisard, O., Boorgula, M.P., Cox, C., Nandy, D., and Seal, S. (2021). Host methylation predicts SARS-CoV-2 infection and clinical outcome. Commun. Med., 1."
},
{
"DOI": "10.1016/j.ebiom.2021.103339",
"doi-asserted-by": "crossref",
"key": "ref_26",
"unstructured": "de Castro Moura, M., Davalos, V., Planas-Serra, L., Alvarez-Errico, D., Arribas, C., Ruiz, M., Aguilera-Albesa, S., Troya, J., Valencia-Ramos, J., and Velez-Santamaria, V. (2021). Epigenome-wide association study of COVID-19 severity with respiratory failure. EBioMedicine, 66."
},
{
"DOI": "10.1186/s13148-021-01102-9",
"doi-asserted-by": "crossref",
"key": "ref_27",
"unstructured": "Balnis, J., Madrid, A., Hogan, K.J., Drake, L.A., Chieng, H.C., Tiwari, A., Vincent, C.E., Chopra, A., Vincent, P.A., and Robek, M.D. (2021). Blood DNA methylation and COVID-19 outcomes. Clin. Epigenet., 13."
},
{
"key": "ref_28",
"unstructured": "NIH (2024, October 30). COVID-19 Treatment Guidelines Panel. Coronavirus Disease 2019. (COVID-19) Treatment Guidelines, Available online: https://www.ncbi.nlm.nih.gov/books/NBK570371/pdf/Bookshelf_NBK570371.pdf."
},
{
"key": "ref_29",
"unstructured": "The R Core Team (2022). R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing."
},
{
"DOI": "10.1093/bioinformatics/btx513",
"article-title": "ChAMP: Updated methylation analysis pipeline for Illumina BeadChips",
"author": "Tian",
"doi-asserted-by": "crossref",
"first-page": "3982",
"journal-title": "Bioinformatics",
"key": "ref_30",
"volume": "33",
"year": "2017"
},
{
"DOI": "10.1093/nar/gkw967",
"doi-asserted-by": "crossref",
"key": "ref_31",
"unstructured": "Zhou, W., Laird, P.W., and Shen, H. (2017). Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res., 45."
},
{
"DOI": "10.1186/gb-2013-14-9-r105",
"doi-asserted-by": "crossref",
"key": "ref_32",
"unstructured": "Nordlund, J., Backlin, C.L., Wahlberg, P., Busche, S., Berglund, E.C., Eloranta, M.L., Flaegstad, T., Forestier, E., Frost, B.M., and Harila-Saari, A. (2013). Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol., 14."
},
{
"DOI": "10.1093/bioinformatics/bts680",
"article-title": "A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data",
"author": "Teschendorff",
"doi-asserted-by": "crossref",
"first-page": "189",
"journal-title": "Bioinformatics",
"key": "ref_33",
"volume": "29",
"year": "2013"
},
{
"DOI": "10.1038/s41467-018-07466-6",
"doi-asserted-by": "crossref",
"key": "ref_34",
"unstructured": "Moss, J., Magenheim, J., Neiman, D., Zemmour, H., Loyfer, N., Korach, A., Samet, Y., Maoz, M., Druid, H., and Arner, P. (2018). Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun., 9."
},
{
"DOI": "10.1093/bioinformatics/btg412",
"article-title": "APE: Analyses of Phylogenetics and Evolution in R language",
"author": "Paradis",
"doi-asserted-by": "crossref",
"first-page": "289",
"journal-title": "Bioinformatics",
"key": "ref_35",
"volume": "20",
"year": "2004"
},
{
"key": "ref_36",
"unstructured": "Kassambara, A., and Mundt, F. (2024, October 30). Factoextra: Extract and Visualize the Results of Multivariate Data Analyses, R Package Version 1.0.7. Available online: https://CRAN.R-project.org/package=factoextra."
},
{
"article-title": "FactoMineR: An R Package for Multivariate Analysis",
"author": "Josse",
"first-page": "1",
"journal-title": "J. Stat. Softw.",
"key": "ref_37",
"volume": "25",
"year": "2008"
},
{
"DOI": "10.1093/bioinformatics/btad184",
"doi-asserted-by": "crossref",
"key": "ref_38",
"unstructured": "Volpe, M., and Das, J. (2023). methylR: A graphical interface for comprehensive DNA methylation array data analysis. Bioinformatics, 39."
},
{
"DOI": "10.1007/978-3-319-24277-4_9",
"doi-asserted-by": "crossref",
"key": "ref_39",
"unstructured": "Wickham, H. (2016). ggplot2: Elegant Graphics for Data Analysis, Springer."
},
{
"DOI": "10.1093/bioinformatics/btw313",
"article-title": "Complex heatmaps reveal patterns and correlations in multidimensional genomic data",
"author": "Gu",
"doi-asserted-by": "crossref",
"first-page": "2847",
"journal-title": "Bioinformatics",
"key": "ref_40",
"volume": "32",
"year": "2016"
},
{
"DOI": "10.1038/s41579-020-00459-7",
"article-title": "Characteristics of SARS-CoV-2 and COVID-19",
"author": "Hu",
"doi-asserted-by": "crossref",
"first-page": "141",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_41",
"volume": "19",
"year": "2021"
},
{
"DOI": "10.1186/gb-2014-15-2-r31",
"doi-asserted-by": "crossref",
"key": "ref_42",
"unstructured": "Jaffe, A.E., and Irizarry, R.A. (2014). Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol., 15."
},
{
"DOI": "10.1093/bioinformatics/btr171",
"article-title": "Independent surrogate variable analysis to deconvolve confounding factors in large-scale microarray profiling studies",
"author": "Teschendorff",
"doi-asserted-by": "crossref",
"first-page": "1496",
"journal-title": "Bioinformatics",
"key": "ref_43",
"volume": "27",
"year": "2011"
},
{
"DOI": "10.1080/01621459.2020.1769635",
"article-title": "Estimating and Accounting for Unobserved Covariates in High-Dimensional Correlated Data",
"author": "McKennan",
"doi-asserted-by": "crossref",
"first-page": "225",
"journal-title": "J. Am. Stat. Assoc.",
"key": "ref_44",
"volume": "117",
"year": "2022"
},
{
"DOI": "10.1186/s40246-024-00578-9",
"doi-asserted-by": "crossref",
"key": "ref_45",
"unstructured": "Ayoub, S.E., Shaker, O.G., Masoud, M., Hassan, E.A., Ezzat, E.M., Ahmed, M.I., Ahmed, R.I., Amin, A.A.I., Abd El Reheem, F., and Khalefa, A.A. (2024). Altered expression of serum lncRNA CASC2 and miRNA-21-5p in COVID-19 patients. Hum. Genom., 18."
},
{
"DOI": "10.1016/j.csbj.2021.01.034",
"article-title": "Chemokines and chemokine receptors during COVID-19 infection",
"author": "Khalil",
"doi-asserted-by": "crossref",
"first-page": "976",
"journal-title": "Comput. Struct. Biotechnol. J.",
"key": "ref_46",
"volume": "19",
"year": "2021"
},
{
"DOI": "10.18632/oncotarget.13562",
"article-title": "Whole genome DNA methylation: Beyond genes silencing",
"author": "Rebbani",
"doi-asserted-by": "crossref",
"first-page": "5629",
"journal-title": "Oncotarget",
"key": "ref_47",
"volume": "8",
"year": "2017"
},
{
"DOI": "10.1038/s41467-019-09234-6",
"doi-asserted-by": "crossref",
"key": "ref_48",
"unstructured": "Zhou, Y., Zhou, B., Pache, L., Chang, M., Khodabakhshi, A.H., Tanaseichuk, O., Benner, C., and Chanda, S.K. (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun., 10."
},
{
"DOI": "10.1038/s41598-022-20062-5",
"doi-asserted-by": "crossref",
"key": "ref_49",
"unstructured": "de Morais Batista, F., Puga, M.A.M., da Silva, P.V., Oliveira, R., Dos Santos, P.C.P., da Silva, B.O., Tatara, M.B., Tsuha, D.H., Dos Santos Pires, M.A., and Goncalves, C.C.M. (2022). Serum biomarkers associated with SARS-CoV-2 severity. Sci. Rep., 12."
},
{
"DOI": "10.1038/s41467-023-40012-7",
"doi-asserted-by": "crossref",
"key": "ref_50",
"unstructured": "Woodruff, M.C., Bonham, K.S., Anam, F.A., Walker, T.A., Faliti, C.E., Ishii, Y., Kaminski, C.Y., Ruunstrom, M.C., Cooper, K.R., and Truong, A.D. (2023). Chronic inflammation, neutrophil activity, and autoreactivity splits long COVID. Nat. Commun., 14."
},
{
"DOI": "10.1155/2019/6273680",
"doi-asserted-by": "crossref",
"key": "ref_51",
"unstructured": "Gonzalez-Jaramillo, V., Portilla-Fernandez, E., Glisic, M., Voortman, T., Ghanbari, M., Bramer, W., Chowdhury, R., Nijsten, T., Dehghan, A., and Franco, O.H. (2019). Epigenetics and Inflammatory Markers: A Systematic Review of the Current Evidence. Int. J. Inflam., 2019."
},
{
"DOI": "10.1186/s13072-021-00428-1",
"doi-asserted-by": "crossref",
"key": "ref_52",
"unstructured": "Konwar, C., Asiimwe, R., Inkster, A.M., Merrill, S.M., Negri, G.L., Aristizabal, M.J., Rider, C.F., MacIsaac, J.L., Carlsten, C., and Kobor, M.S. (2021). Risk-focused differences in molecular processes implicated in SARS-CoV-2 infection: Corollaries in DNA methylation and gene expression. Epigenetics Chromatin, 14."
},
{
"DOI": "10.1164/rccm.201207-1280OC",
"article-title": "Systemic steroid exposure is associated with differential methylation in chronic obstructive pulmonary disease",
"author": "Wan",
"doi-asserted-by": "crossref",
"first-page": "1248",
"journal-title": "Am. J. Respir. Crit. Care Med.",
"key": "ref_53",
"volume": "186",
"year": "2012"
},
{
"DOI": "10.1038/s41598-021-95532-3",
"doi-asserted-by": "crossref",
"key": "ref_54",
"unstructured": "Winkley, K., Banerjee, D., Bradley, T., Koseva, B., Cheung, W.A., Selvarangan, R., Pastinen, T., and Grundberg, E. (2021). Immune cell residency in the nasal mucosa may partially explain respiratory disease severity across the age range. Sci. Rep., 11."
},
{
"DOI": "10.1038/s41385-020-00359-2",
"article-title": "The central role of the nasal microenvironment in the transmission, modulation, and clinical progression of SARS-CoV-2 infection",
"author": "Gallo",
"doi-asserted-by": "crossref",
"first-page": "305",
"journal-title": "Mucosal Immunol.",
"key": "ref_55",
"volume": "14",
"year": "2021"
},
{
"DOI": "10.1165/rcmb.2022-0433LE",
"article-title": "Whole-Genome Methylation Sequencing Reveals that COVID-19-induced Epigenetic Dysregulation Remains 1 Year after Hospital Discharge",
"author": "Balnis",
"doi-asserted-by": "crossref",
"first-page": "594",
"journal-title": "Am. J. Respir. Cell Mol. Biol.",
"key": "ref_56",
"volume": "68",
"year": "2023"
},
{
"DOI": "10.1080/15592294.2019.1638701",
"article-title": "DNA hypermethylation in disease: Mechanisms and clinical relevance",
"author": "Ehrlich",
"doi-asserted-by": "crossref",
"first-page": "1141",
"journal-title": "Epigenetics",
"key": "ref_57",
"volume": "14",
"year": "2019"
},
{
"DOI": "10.1002/JLB.5HI0720-466R",
"article-title": "Genome-wide DNA methylation profiling of peripheral blood reveals an epigenetic signature associated with severe COVID-19",
"author": "Corley",
"doi-asserted-by": "crossref",
"first-page": "21",
"journal-title": "J. Leukoc. Biol.",
"key": "ref_58",
"volume": "110",
"year": "2021"
},
{
"DOI": "10.3390/v15020553",
"doi-asserted-by": "crossref",
"key": "ref_59",
"unstructured": "Silva, M.J.A., Ribeiro, L.R., Gouveia, M.I.M., Marcelino, B.D.R., Santos, C.S.D., Lima, K.V.B., and Lima, L. (2023). Hyperinflammatory Response in COVID-19: A Systematic Review. Viruses, 15."
},
{
"DOI": "10.1016/j.cell.2020.08.001",
"article-title": "Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment",
"author": "Reusch",
"doi-asserted-by": "crossref",
"first-page": "1419",
"journal-title": "Cell",
"key": "ref_60",
"volume": "182",
"year": "2020"
},
{
"DOI": "10.1038/s41418-021-00805-z",
"article-title": "Patients with COVID-19: In the dark-NETs of neutrophils",
"author": "Ackermann",
"doi-asserted-by": "crossref",
"first-page": "3125",
"journal-title": "Cell Death Differ.",
"key": "ref_61",
"volume": "28",
"year": "2021"
},
{
"DOI": "10.1038/s41467-022-34910-5",
"doi-asserted-by": "crossref",
"key": "ref_62",
"unstructured": "Biering, S.B., de Gomes Sousa, F.T., Tjang, L.V., Pahmeier, F., Zhu, C., Ruan, R., Blanc, S.F., Patel, T.S., Worthington, C.M., and Glasner, D.R. (2022). SARS-CoV-2 Spike triggers barrier dysfunction and vascular leak via integrins and TGF-β signaling. Nat. Commun., 13."
},
{
"DOI": "10.1016/j.bcp.2021.114812",
"doi-asserted-by": "crossref",
"key": "ref_63",
"unstructured": "Giacomelli, C., Piccarducci, R., Marchetti, L., Romei, C., and Martini, C. (2021). Pulmonary fibrosis from molecular mechanisms to therapeutic interventions: Lessons from post-COVID-19 patients. Biochem. Pharmacol., 193."
},
{
"DOI": "10.3390/ph14070695",
"doi-asserted-by": "crossref",
"key": "ref_64",
"unstructured": "Vassiliou, A.G., Zacharis, A., Keskinidou, C., Jahaj, E., Pratikaki, M., Gallos, P., Dimopoulou, I., Kotanidou, A., and Orfanos, S.E. (2021). Soluble Angiotensin Converting Enzyme 2 (ACE2) Is Upregulated and Soluble Endothelial Nitric Oxide Synthase (eNOS) Is Downregulated in COVID-19-induced Acute Respiratory Distress Syndrome (ARDS). Pharmaceuticals, 14."
},
{
"DOI": "10.1016/j.niox.2021.04.003",
"article-title": "Implications of SARS-CoV-2 infection on eNOS and iNOS activity: Consequences for the respiratory and vascular systems",
"author": "Guimaraes",
"doi-asserted-by": "crossref",
"first-page": "64",
"journal-title": "Nitric Oxide",
"key": "ref_65",
"volume": "111–112",
"year": "2021"
},
{
"DOI": "10.1038/s41440-023-01460-2",
"article-title": "The role of SARS-CoV-2-mediated NF-kappaB activation in COVID-19 patients",
"author": "Zhou",
"doi-asserted-by": "crossref",
"first-page": "375",
"journal-title": "Hypertens. Res.",
"key": "ref_66",
"volume": "47",
"year": "2024"
},
{
"DOI": "10.1016/j.virusres.2023.199086",
"doi-asserted-by": "crossref",
"key": "ref_67",
"unstructured": "Nie, Y., Mou, L., Long, Q., Deng, D., Hu, R., Cheng, J., and Wu, J. (2023). SARS-CoV-2 ORF3a positively regulates NF-kappaB activity by enhancing IKKbeta-NEMO interaction. Virus Res., 328."
},
{
"DOI": "10.1371/journal.pone.0274841",
"doi-asserted-by": "crossref",
"key": "ref_68",
"unstructured": "Sharif-Askari, F.S., Sharif-Askari, N.S., Hafezi, S., Mdkhana, B., Alsayed, H.A.H., Ansari, A.W., Mahboub, B., Zakeri, A.M., Temsah, M.H., and Zahir, W. (2022). Interleukin-17, a salivary biomarker for COVID-19 severity. PLoS ONE, 17."
},
{
"DOI": "10.3389/fimmu.2022.1020624",
"doi-asserted-by": "crossref",
"key": "ref_69",
"unstructured": "Al-Qahtani, A.A., Pantazi, I., Alhamlan, F.S., Alothaid, H., Matou-Nasri, S., Sourvinos, G., Vergadi, E., and Tsatsanis, C. (2022). SARS-CoV-2 modulates inflammatory responses of alveolar epithelial type II cells via PI3K/AKT pathway. Front. Immunol., 13."
},
{
"DOI": "10.1007/s00018-021-04085-1",
"article-title": "The MEK1/2-inhibitor ATR-002 efficiently blocks SARS-CoV-2 propagation and alleviates pro-inflammatory cytokine/chemokine responses",
"author": "Schreiber",
"doi-asserted-by": "crossref",
"first-page": "65",
"journal-title": "Cell Mol. Life Sci.",
"key": "ref_70",
"volume": "79",
"year": "2022"
},
{
"DOI": "10.1128/mbio.01007-23",
"doi-asserted-by": "crossref",
"key": "ref_71",
"unstructured": "Higgins, C.A., Nilsson-Payant, B.E., Bonaventure, B., Kurland, A.P., Ye, C., Yaron, T.M., Johnson, J.L., Adhikary, P., Golynker, I., and Panis, M. (2023). SARS-CoV-2 hijacks p38beta/MAPK11 to promote virus replication. mBio, 14."
},
{
"DOI": "10.1001/jamacardio.2021.5133",
"article-title": "Association of Complement and MAPK Activation With SARS-CoV-2-Associated Myocardial Inflammation",
"author": "Weckbach",
"doi-asserted-by": "crossref",
"first-page": "286",
"journal-title": "JAMA Cardiol.",
"key": "ref_72",
"volume": "7",
"year": "2022"
},
{
"DOI": "10.1016/j.cell.2023.07.019",
"article-title": "Epigenetic memory of coronavirus infection in innate immune cells and their progenitors",
"author": "Cheong",
"doi-asserted-by": "crossref",
"first-page": "3882",
"journal-title": "Cell",
"key": "ref_73",
"volume": "186",
"year": "2023"
},
{
"DOI": "10.1016/j.it.2020.10.012",
"article-title": "A Potential Role of Interleukin 10 in COVID-19 Pathogenesis",
"author": "Lu",
"doi-asserted-by": "crossref",
"first-page": "3",
"journal-title": "Trends Immunol.",
"key": "ref_74",
"volume": "42",
"year": "2021"
},
{
"DOI": "10.1016/j.ebiom.2023.104552",
"doi-asserted-by": "crossref",
"key": "ref_75",
"unstructured": "Espin, E., Yang, C., Shannon, C.P., Assadian, S., He, D., and Tebbutt, S.J. (2023). Cellular and molecular biomarkers of long COVID: A scoping review. EBioMedicine, 91."
},
{
"DOI": "10.1016/j.cell.2020.10.004",
"article-title": "SARS-CoV-2 Disrupts Splicing, Translation, and Protein Trafficking to Suppress Host Defenses",
"author": "Banerjee",
"doi-asserted-by": "crossref",
"first-page": "1325",
"journal-title": "Cell",
"key": "ref_76",
"volume": "183",
"year": "2020"
},
{
"DOI": "10.2217/fvl-2018-0008",
"article-title": "Post-translational modifications of coronavirus proteins: Roles and function",
"author": "Fung",
"doi-asserted-by": "crossref",
"first-page": "405",
"journal-title": "Future Virol.",
"key": "ref_77",
"volume": "13",
"year": "2018"
},
{
"DOI": "10.1038/s41591-020-0944-y",
"article-title": "A single-cell atlas of the peripheral immune response in patients with severe COVID-19",
"author": "Wilk",
"doi-asserted-by": "crossref",
"first-page": "1070",
"journal-title": "Nat. Med.",
"key": "ref_78",
"volume": "26",
"year": "2020"
},
{
"DOI": "10.1128/JVI.00985-20",
"doi-asserted-by": "crossref",
"key": "ref_79",
"unstructured": "Vanderheiden, A., Ralfs, P., Chirkova, T., Upadhyay, A.A., Zimmerman, M.G., Bedoya, S., Aoued, H., Tharp, G.M., Pellegrini, K.L., and Manfredi, C. (2020). Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures. J. Virol., 94."
},
{
"DOI": "10.1016/j.cell.2020.04.026",
"article-title": "Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19",
"author": "Liu",
"doi-asserted-by": "crossref",
"first-page": "1036",
"journal-title": "Cell",
"key": "ref_80",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1038/s41586-022-05282-z",
"article-title": "SARS-CoV-2 disrupts host epigenetic regulation via histone mimicry",
"author": "Kee",
"doi-asserted-by": "crossref",
"first-page": "381",
"journal-title": "Nature",
"key": "ref_81",
"volume": "610",
"year": "2022"
},
{
"DOI": "10.1016/j.bbadis.2023.166836",
"doi-asserted-by": "crossref",
"key": "ref_82",
"unstructured": "Saksena, N.K., Reddy, S.B., Miranda-Saksena, M., Cardoso, T.H.S., Silva, E.M.A., Ferreira, J.C., and Rabeh, W.M. (2023). SARS-CoV-2 variants, its recombinants and epigenomic exploitation of host defenses. Biochim. Et. Biophys. Acta (BBA)—Mol. Basis Dis., 1869."
},
{
"DOI": "10.1016/j.clim.2020.108410",
"doi-asserted-by": "crossref",
"key": "ref_83",
"unstructured": "Sawalha, A.H., Zhao, M., Coit, P., and Lu, Q. (2020). Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients. Clin. Immunol., 215."
},
{
"DOI": "10.3389/fmed.2021.685333",
"doi-asserted-by": "crossref",
"key": "ref_84",
"unstructured": "Falcao-Holanda, R.B., Brunialti, M.K.C., Jasiulionis, M.G., and Salomao, R. (2021). Epigenetic Regulation in Sepsis, Role in Pathophysiology and Therapeutic Perspective. Front. Med., 8."
}
],
"reference-count": 84,
"references-count": 84,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/2073-4409/14/21/1673"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Altered DNA Methylation Pattern Contributes to Differential Epigenetic Immune Signaling in the Upper Respiratory Airway of Unvaccinated COVID-19 Patients",
"type": "journal-article",
"update-policy": "https://doi.org/10.3390/mdpi_crossmark_policy",
"volume": "14"
}