SARS-CoV-2 infection induces DNA damage, through CHK1 degradation and impaired 53BP1 recruitment, and cellular senescence
et al., Nature Cell Biology, doi:10.1038/s41556-023-01096-x, Mar 2023
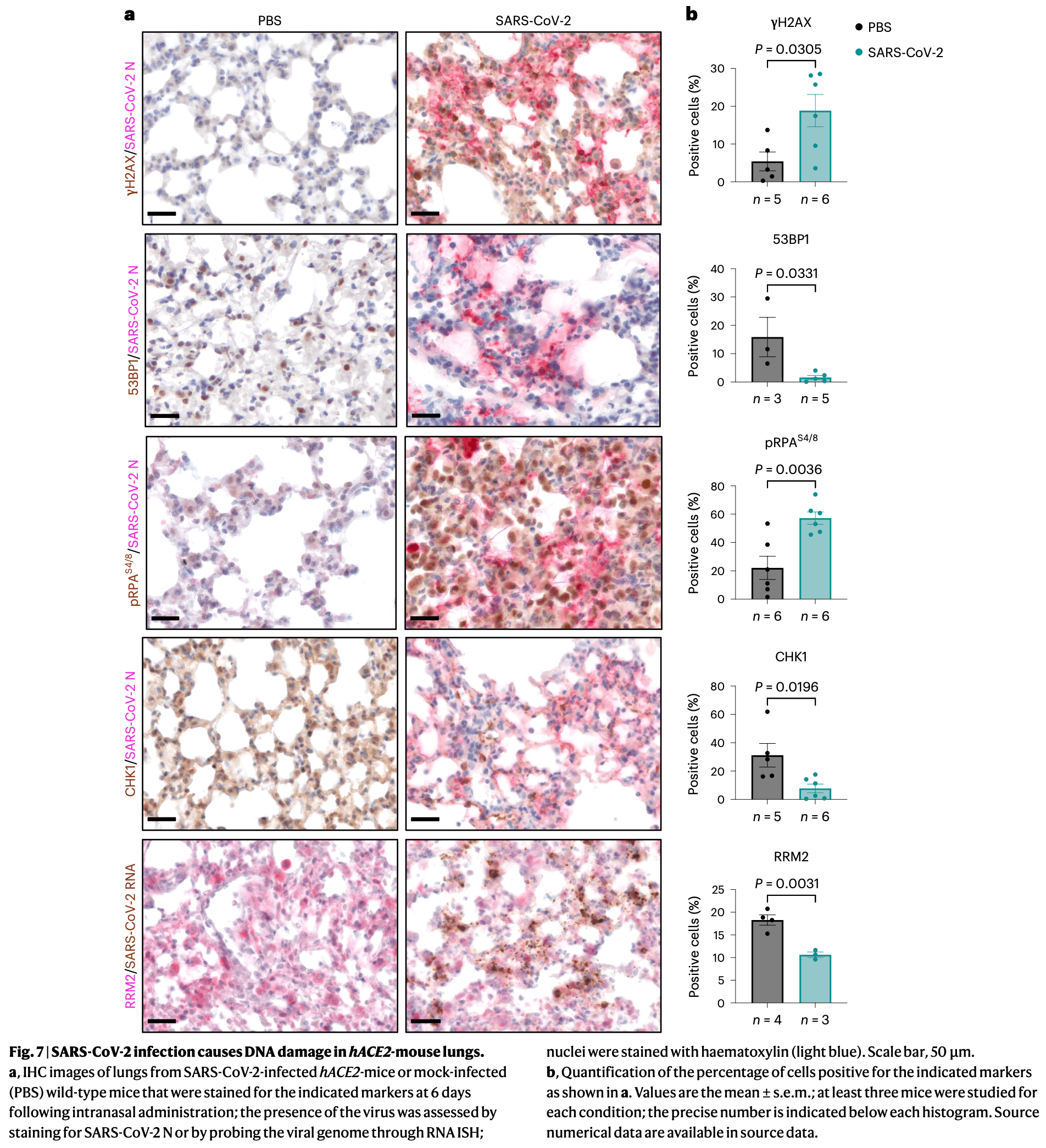
In vitro, animal, and ex vivo study showing that SARS-CoV-2 causes DNA damage through multiple mechanisms. Using cultured human cells (Huh7, Calu-3, and primary nasal epithelial cells), infected mice, and tissue samples from COVID-19 patients, authors found that viral proteins ORF6 and NSP13 degrade the DNA repair protein CHK1 through proteasome and autophagy pathways respectively, leading to deoxynucleoside triphosphate (dNTP) shortage, impaired DNA replication, and cellular senescence. Additionally, the viral N-protein interferes with DNA repair by competing with 53BP1 protein for damage-induced long non-coding RNAs. DNA damage was confirmed in lung tissues from infected mice and COVID-19 patients.
Gioia et al., 9 Mar 2023, peer-reviewed, 29 authors.
Contact: fabrizio.dadda@ifom.eu.
Ex vivo studies are an important part of preclinical research, however results may be very different in vivo.
SARS-CoV-2 infection induces DNA damage, through CHK1 degradation and impaired 53BP1 recruitment, and cellular senescence
Nature Cell Biology, doi:10.1038/s41556-023-01096-x
Gating strategy their SSC-A vs. FSC-A and SSC-A vs. SSC-H parameters. 561 nm laser and 695/40 filter were used for propidium iodide detection; 488 nm laser and 530/30 filter were used for BrdU and CHK1 detection. Tick this box to confirm that a figure exemplifying the gating strategy is provided in the Supplementary Information.
Publisher's note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons. org/licenses/by/4.0/ . © The Author(s) 2023 1 IFOM ETS -The AIRC Institute of Molecular Oncology, Milan, Italy. 2 International Centre for Genetic Engineering and Biotechnology, Trieste, Italy. 3 University of Palermo, Palermo, Italy. 4 IRCCS San Raffaele Scientific Institute & University, Milan, Italy. 5 University of Trieste, Trieste, Italy. 6 University of Padova, Padova, Italy. 7 Fondazione IRCCS Istituto Neurologico Carlo Besta, Milan, Italy. 8 Cogentech Società Benefit srl, Milan, Italy. 9 Institute of..
References
Addetia, SARS-CoV-2 orf6 disrupts bidirectional nucleocytoplasmic transport through interactions with rae1 and nup98, mBio
Ahmadi, Moradi, In silico analysis suggests the RNAi-enhancing antibiotic enoxacin as a potential inhibitor of SARS-CoV-2 infection, Sci. Rep
Aird, Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence, Cell Rep
Anacker, HPV31 utilizes the ATR-Chk1 pathway to maintain elevated RRM2 levels and a replication-competent environment in differentiating keratinocytes, Virology
Banerjee, SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses, Cell
Bekker-Jensen, Lukas, Melander, Bartek, Lukas, Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1, J. Cell Biol
Bestle, TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells, Life Sci. Alliance
Blanco-Melo, Imbalanced host response to SARS-CoV-2 drives development of COVID-19, Cell
Bouhaddou, The global phosphorylation landscape of SARS-CoV-2 infection, Cell
Bussani, Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology, EBioMedicine
Cao, Accelerated biological aging in COVID-19 patients, Nat. Commun
Chen, Liquid-liquid phase separation by SARS-CoV-2 nucleocapsid protein and RNA, Cell Res, doi:10.1038/s41422-020-00408-2
Choi, Bowman, Jung, Autophagy during viral infection-a double-edged sword, Nat. Rev. Microbiol
D'agnillo, Lung epithelial and endothelial damage, loss of tissue repair, inhibition of fibrinolysis, and cellular senescence in fatal COVID-19, Sci. Transl. Med
D'alessandro, BRCA2 controls DNA:RNA hybrid level at DSBs by mediating RNase H2 recruitment, Nat. Commun
Di Fagagna, Living on a break: cellular senescence as a DNA-damage response, Nat. Rev. Cancer
Di Micco, Krizhanovsky, Baker, Di Fagagna, Cellular senescence in ageing: from mechanisms to therapeutic opportunities, Nat. Rev. Mol. Cell Biol
Dimri, A biomarker that identifies senescent human cells in culture and in aging skin in vivo, Proc. Natl Acad. Sci
Evangelou, Pulmonary infection by SARS-CoV-2 induces senescence accompanied by an inflammatory phenotype in severe COVID-19: possible implications for viral mutagenesis, Eur. Respir. J, doi:10.1183/13993003.02951-2021
Ferraro, Franzolin, Pontarin, Reichard, Bianchi, Quantitation of cellular deoxynucleoside triphosphates, Nature Cell Biology
Francia, Site-specific DICER and DROSHA RNA products control the DNA-damage response, Nature, doi:10.1038/nature11179
Frangini, Synthesis of mitochondrial DNA precursors during myogenesis, an analysis in purified C2C12 myotubes, J. Biol. Chem
Fumagalli, Administration of aerosolized SARS-CoV-2 to K18-hACE2 mice uncouples respiratory infection from fatal neuroinvasion, Sci. Immunol
Fumagalli, Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation, Nat. Cell Biol
Garcia, Antiviral drug screen identifies DNA-damage response inhibitor as potent blocker of SARS-CoV-2 replication, Cell Rep
Gioia, Pharmacological boost of DNA damage response and repair by enhanced biogenesis of DNA damage response RNAs, Sci. Rep
Gong, ATR-CHK1-E2F3 signaling transactivates human ribonucleotide reductase small subunit M2 for DNA repair induced by the chemical carcinogen MNNG, Biochim. Biophys. Acta Gene Regul. Mech
González Besteiro, Chk1 loss creates replication barriers that compromise cell survival independently of excess origin firing, EMBO J
Gordon, A SARS-CoV-2 protein interaction map reveals targets for drug repurposing, Nature
Green, Levine, To be or not to be? How selective autophagy and cell death govern cell fate, Cell
Gunn, Stark, I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks, Methods Mol. Biol
Gussow, Genomic determinants of pathogenicity in SARS-CoV-2 and other human coronaviruses, Proc. Natl Acad. Sci. USA
Harding, Mitotic progression following DNA damage enables pattern recognition within micronuclei, Nature
Higa, Mihaylov, Banks, Zheng, Zhang, Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint, Nat. Cell Biol
Hu, Mccall, Ohta, Xiong, Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage, Nat. Cell Biol
Jackson, Bartek, The DNA-damage response in human biology and disease, Nature
Jiang, Mei, SARS-CoV -2 spike impairs DNA damage repair and inhibits V(D)J recombination in vitro, Viruses, doi:10.3390/v13102056
Kilic, Phase separation of 53 BP 1 determines liquid-like behavior of DNA repair compartments, EMBO J, doi:10.15252/embj.2018101379
Kim, The architecture of SARS-CoV-2 transcriptome, Cell
Klionsky, Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition), Autophagy
Koç, Wheeler, Mathews, Merrill, Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools, J. Biol. Chem
Kratzel, A genome-wide CRISPR screen identifies interactors of the autophagy pathway as conserved coronavirus targets, PLoS Biol
Lee, Virus-induced senescence is driver and therapeutic target in COVID-19, Nature
Li, Chen, The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer, J. Exp. Med
Li, SARS-CoV-2 induces double-stranded RNA-mediated innate immune responses in respiratory epithelial-derived cells and cardiomyocytes, Proc. Natl Acad. Sci. USA
Licastro, Isolation and full-length genome characterization of SARS-CoV-2 from COVID-19 cases in Northern Italy, J. Virol
Lilley, A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses, EMBO J
Lilley, Schwartz, Weitzman, Using or abusing: viruses and the cellular DNA damage response, Trends Microbiol
Lipskaia, Evidence that SARS-CoV-2 induces lung-cell senescence: potential impact on COVID-19 lung disease, Am. J. Respir. Cell Mol. Biol, doi:10.1165/rcmb.2021-0205le
Liu, SARS-CoV-2-host proteome interactions for antiviral drug discovery, Mol. Syst. Biol
Luo, Interplay between the virus and the ubiquitin-proteasome system: molecular mechanism of viral pathogenesis, Curr. Opin. Virol
Ma-Lauer, P53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1, Proc. Natl Acad. Sci. USA
Mavrikaki, Lee, Solomon, Slack, Severe COVID-19 is associated with molecular signatures of aging in the human brain, Nat. Aging
Mehta, COVID-19: consider cytokine storm syndromes and immunosuppression, Lancet
Michelini, Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks, Nat. Cell Biol
Miorin, SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling, Proc. Natl Acad. Sci. USA
Morello, T cells expressing receptor recombination/ revision machinery are detected in the tumor microenvironment and expanded in genomically over-unstable models, Cancer Immunol. Res
Nalbandian, Post-acute COVID-19 syndrome, Nat. Med
Naruyama, Essential role of Chk1 in S phase progression through regulation of RNR2 expression, Biochem. Biophys. Res. Commun
Perdikari, SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs, EMBO J, doi:10.15252/embj.2020106478
Pessina, Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors, Nat. Cell Biol
Polo, Jackson, Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications, Genes Dev
Pontarin, Ferraro, Bee, Reichard, Bianchi, Mammalian ribonucleotide reductase subunit p53R2 is required for mitochondrial DNA replication and DNA repair in quiescent cells, Proc. Natl Acad. Sci. USA
Ren, Micronucleus production, activation of DNA damage response and cGAS-STING signaling in syncytia induced by SARS-CoV-2 infection, Biol. Direct
Rodier, Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion, Nat. Cell Biol
Rossiello, Herbig, Longhese, Fumagalli, Di Fagagna, Irreparable telomeric DNA damage and persistent DDR signalling as a shared causative mechanism of cellular senescence and ageing, Curr. Opin. Genet. Dev
Rowland, Intracellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein: absence of nucleolar accumulation during infection and after expression as a recombinant protein in Vero cells, J. Virol
Ryan, Hollingworth, Grand, Activation of the DNA damage response by RNA viruses, Biomolecules
Sansam, DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint, Genes Dev
Savastano, De Opakua, Rankovic, Zweckstetter, Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates, Nat. Commun, doi:10.1101/2020.06.18.160648
Schmidt, The SARS-CoV-2 RNA-protein interactome in infected human cells, Nat. Microbiol, doi:10.1038/s41564-020-00846-z
Schmitt, COVID-19 and cellular senescence, Nat. Rev. Immunol, doi:10.1038/s41577-022-00785-2
Sepe, Materials & experimental systems n/a Involved in the study Antibodies Eukaryotic cell lines Palaeontology and archaeology Animals and other organisms Clinical data Dual use research of concern Methods n/a Involved in the study ChIP-seq Flow cytometry MRI-based neuroimaging Antibodies Antibodies used gH2AX (Ser139) Abcam ab11174 gH2AX (Ser139) Millipore 05-636 53BP1 Bethyl A303-906A 53BP1 Novus NB100-304 ACE2 Abcam ab15348 ATM Abcam ab32420 ATR Santa Cruz sc-1887 Beclin 1 Bethyl A302-566A-T Beta-actin Sigma-Aldrich A2228 BrdU BDbioscience 347580 CD68 Abcam ab125212 CDT1 Cell Signaling #8064 cGAS Cell Signaling #15102 CHK1 Novus NB100-46 CHK1 (2G1D5) Cell Signaling #2360 CHK1 (ST57-09) ThermoFisher MA532180 CHK2 Millipore 05-649 Cleaved Caspase-3 (Asp 175) Cell Signaling 9661 DNA-PK Abcam ab32566 HA-tag Abcam ab236632 Histone H3 Abcam ab10799 Human Cytokeratin 8/18 (EP17/EP30) Dako M3652 ISceI (FL-86, doi:10.15252/embr.202153658
Shamanna, WRN regulates pathway choice between classical and alternative non-homologous end joining, Nat. Commun
Shechter, Costanzo, Gautier, Regulation of DNA replication by ATR: signaling in response to DNA intermediates, DNA Repair
Sui, SARS-CoV-2 NSP13 inhibits type I IFN production by degradation of TBK1 via p62-dependent selective autophagy, J. Immunol
Thomsen, Lack of immunological DNA sensing in hepatocytes facilitates hepatitis B virus infection, Hepatology
Timani, Nuclear/nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus, Virus Res
Trimarchi, Lees, Sibling rivalry in the E2F family, Nat. Rev. Mol. Cell Biol
Tsuji, SARS-CoV-2 infection triggers paracrine senescence and leads to a sustained senescence-associated inflammatory response, Nat. Aging
Turnell, Grand, DNA viruses and the cellular DNA-damage response, J. Gen. Virol
V'kovski, Disparate temperature-dependent virus-host dynamics for SARS-CoV-2 and SARS-CoV in the human respiratory epithelium, PLoS Biol
V'kovski, Kratzel, Steiner, Stalder, Thiel, Coronavirus biology and replication: implications for SARS-CoV-2, Nat. Rev. Microbiol
Victor, SARS-CoV-2 triggers DNA damage response in Vero E6 cells, Biochem. Biophys. Res. Commun
Wang, Han, Feng, Wang, Zhang, Coupling cellular localization and function of checkpoint kinase 1 (Chk1) in checkpoints and cell viability, J. Biol. Chem
Wang, Shi, Xu, Yin, SARS-CoV-2 nucleocapsid protein undergoes liquid-liquid phase separation into stress granules through its N-terminal intrinsically disordered region, Cell Discov
Wang, Targeting liquid-liquid phase separation of SARS-CoV-2 nucleocapsid protein promotes innate antiviral immunity by elevating MAVS activity, Nat. Cell Biol
Weitzman, Lilley, Chaurushiya, Genomes in conflict: maintaining genome integrity during virus infection, Annu. Rev. Microbiol
Yang, Rao, Structural biology of SARS-CoV-2 and implications for therapeutic development, Nat. Rev. Microbiol
Zhang, Jones, Martin, Caplen, Pommier, Implication of checkpoint kinase-dependent up-regulation of ribonucleotide reductase R2 in DNA damage response, J. Biol. Chem
Zhou, Sensing of cytoplasmic chromatin by cGAS activates innate immune response in SARS-CoV-2 infection, Signal Transduct. Target. Ther, doi:10.1038/s41392-021-00800-3
Zimmermann, De Lange, 53BP1: pro choice in DNA repair, Trends Cell Biol
DOI record:
{
"DOI": "10.1038/s41556-023-01096-x",
"ISSN": [
"1465-7392",
"1476-4679"
],
"URL": "http://dx.doi.org/10.1038/s41556-023-01096-x",
"abstract": "<jats:title>Abstract</jats:title><jats:p>Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the RNA virus responsible for the coronavirus disease 2019 (COVID-19) pandemic. Although SARS-CoV-2 was reported to alter several cellular pathways, its impact on DNA integrity and the mechanisms involved remain unknown. Here we show that SARS-CoV-2 causes DNA damage and elicits an altered DNA damage response. Mechanistically, SARS-CoV-2 proteins ORF6 and NSP13 cause degradation of the DNA damage response kinase CHK1 through proteasome and autophagy, respectively. CHK1 loss leads to deoxynucleoside triphosphate (dNTP) shortage, causing impaired S-phase progression, DNA damage, pro-inflammatory pathways activation and cellular senescence. Supplementation of deoxynucleosides reduces that. Furthermore, SARS-CoV-2 N-protein impairs 53BP1 focal recruitment by interfering with damage-induced long non-coding RNAs, thus reducing DNA repair. Key observations are recapitulated in SARS-CoV-2-infected mice and patients with COVID-19. We propose that SARS-CoV-2, by boosting ribonucleoside triphosphate levels to promote its replication at the expense of dNTPs and by hijacking damage-induced long non-coding RNAs’ biology, threatens genome integrity and causes altered DNA damage response activation, induction of inflammation and cellular senescence.</jats:p>",
"alternative-id": [
"1096"
],
"assertion": [
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Received",
"name": "received",
"order": 1,
"value": "17 December 2021"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "25 January 2023"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "First Online",
"name": "first_online",
"order": 3,
"value": "9 March 2023"
},
{
"group": {
"label": "Competing interests",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 1,
"value": "M.I. participates in advisory boards/consultancies for Gilead Sciences, Roche, Third Rock Ventures, Antios Therapeutics, Asher Bio, Amgen, Allovir. All the other authors declare no competing interests."
}
],
"author": [
{
"ORCID": "https://orcid.org/0000-0002-4993-5283",
"affiliation": [],
"authenticated-orcid": false,
"family": "Gioia",
"given": "Ubaldo",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0000-0003-4627-1589",
"affiliation": [],
"authenticated-orcid": false,
"family": "Tavella",
"given": "Sara",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Martínez-Orellana",
"given": "Pamela",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Cicio",
"given": "Giada",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Colliva",
"given": "Andrea",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ceccon",
"given": "Marta",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Cabrini",
"given": "Matteo",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Henriques",
"given": "Ana C.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-2583-2498",
"affiliation": [],
"authenticated-orcid": false,
"family": "Fumagalli",
"given": "Valeria",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Paldino",
"given": "Alessia",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-8446-0105",
"affiliation": [],
"authenticated-orcid": false,
"family": "Presot",
"given": "Ettore",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Rajasekharan",
"given": "Sreejith",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Iacomino",
"given": "Nicola",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Pisati",
"given": "Federica",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Matti",
"given": "Valentina",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Sepe",
"given": "Sara",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Conte",
"given": "Matilde I.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Barozzi",
"given": "Sara",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-7247-8084",
"affiliation": [],
"authenticated-orcid": false,
"family": "Lavagnino",
"given": "Zeno",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-2510-2399",
"affiliation": [],
"authenticated-orcid": false,
"family": "Carletti",
"given": "Tea",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Volpe",
"given": "Maria Concetta",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Cavalcante",
"given": "Paola",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-9370-2671",
"affiliation": [],
"authenticated-orcid": false,
"family": "Iannacone",
"given": "Matteo",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-1092-2653",
"affiliation": [],
"authenticated-orcid": false,
"family": "Rampazzo",
"given": "Chiara",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Bussani",
"given": "Rossana",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-0821-6231",
"affiliation": [],
"authenticated-orcid": false,
"family": "Tripodo",
"given": "Claudio",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-6705-3076",
"affiliation": [],
"authenticated-orcid": false,
"family": "Zacchigna",
"given": "Serena",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-8903-8202",
"affiliation": [],
"authenticated-orcid": false,
"family": "Marcello",
"given": "Alessandro",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-9603-5966",
"affiliation": [],
"authenticated-orcid": false,
"family": "d’Adda di Fagagna",
"given": "Fabrizio",
"sequence": "additional"
}
],
"container-title": "Nature Cell Biology",
"container-title-short": "Nat Cell Biol",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"link.springer.com"
]
},
"created": {
"date-parts": [
[
2023,
3,
9
]
],
"date-time": "2023-03-09T17:03:17Z",
"timestamp": 1678381397000
},
"deposited": {
"date-parts": [
[
2023,
4,
26
]
],
"date-time": "2023-04-26T12:09:15Z",
"timestamp": 1682510955000
},
"indexed": {
"date-parts": [
[
2025,
8,
20
]
],
"date-time": "2025-08-20T12:28:10Z",
"timestamp": 1755692890865,
"version": "3.37.3"
},
"is-referenced-by-count": 85,
"issue": "4",
"issued": {
"date-parts": [
[
2023,
3,
9
]
]
},
"journal-issue": {
"issue": "4",
"published-print": {
"date-parts": [
[
2023,
4
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
3,
9
]
],
"date-time": "2023-03-09T00:00:00Z",
"timestamp": 1678320000000
}
},
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
3,
9
]
],
"date-time": "2023-03-09T00:00:00Z",
"timestamp": 1678320000000
}
}
],
"link": [
{
"URL": "https://www.nature.com/articles/s41556-023-01096-x.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41556-023-01096-x",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41556-023-01096-x.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"page": "550-564",
"prefix": "10.1038",
"published": {
"date-parts": [
[
2023,
3,
9
]
]
},
"published-online": {
"date-parts": [
[
2023,
3,
9
]
]
},
"published-print": {
"date-parts": [
[
2023,
4
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference": [
{
"DOI": "10.1038/s41579-020-00468-6",
"author": "P V’kovski",
"doi-asserted-by": "publisher",
"first-page": "155",
"journal-title": "Nat. Rev. Microbiol.",
"key": "1096_CR1",
"unstructured": "V’kovski, P., Kratzel, A., Steiner, S., Stalder, H. & Thiel, V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 19, 155–170 (2021).",
"volume": "19",
"year": "2021"
},
{
"DOI": "10.1038/s41579-021-00630-8",
"author": "H Yang",
"doi-asserted-by": "publisher",
"first-page": "685",
"journal-title": "Nat. Rev. Microbiol.",
"key": "1096_CR2",
"unstructured": "Yang, H. & Rao, Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 19, 685–700 (2021).",
"volume": "19",
"year": "2021"
},
{
"DOI": "10.1038/s41579-018-0003-6",
"author": "Y Choi",
"doi-asserted-by": "publisher",
"first-page": "341",
"journal-title": "Nat. Rev. Microbiol.",
"key": "1096_CR3",
"unstructured": "Choi, Y., Bowman, J. W. & Jung, J. U. Autophagy during viral infection—a double-edged sword. Nat. Rev. Microbiol. 16, 341–354 (2018).",
"volume": "16",
"year": "2018"
},
{
"DOI": "10.1016/j.coviro.2015.09.005",
"author": "H Luo",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Curr. Opin. Virol.",
"key": "1096_CR4",
"unstructured": "Luo, H. Interplay between the virus and the ubiquitin–proteasome system: molecular mechanism of viral pathogenesis. Curr. Opin. Virol. 17, 1–10 (2016).",
"volume": "17",
"year": "2016"
},
{
"DOI": "10.1016/j.tim.2007.01.003",
"author": "CE Lilley",
"doi-asserted-by": "publisher",
"first-page": "119",
"journal-title": "Trends Microbiol.",
"key": "1096_CR5",
"unstructured": "Lilley, C. E., Schwartz, R. A. & Weitzman, M. D. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 15, 119–126 (2007).",
"volume": "15",
"year": "2007"
},
{
"DOI": "10.3390/biom6010002",
"author": "EL Ryan",
"doi-asserted-by": "publisher",
"first-page": "2",
"journal-title": "Biomolecules",
"key": "1096_CR6",
"unstructured": "Ryan, E. L., Hollingworth, R. & Grand, R. J. Activation of the DNA damage response by RNA viruses. Biomolecules 6, 2–24 (2016).",
"volume": "6",
"year": "2016"
},
{
"DOI": "10.1016/j.cell.2020.06.034",
"author": "M Bouhaddou",
"doi-asserted-by": "publisher",
"first-page": "685",
"journal-title": "Cell",
"key": "1096_CR7",
"unstructured": "Bouhaddou, M. et al. The global phosphorylation landscape of SARS-CoV-2 infection. Cell 182, 685–712.e19 (2020).",
"volume": "182",
"year": "2020"
},
{
"DOI": "10.1016/j.bbrc.2021.09.024",
"author": "J Victor",
"doi-asserted-by": "publisher",
"first-page": "141",
"journal-title": "Biochem. Biophys. Res. Commun.",
"key": "1096_CR8",
"unstructured": "Victor, J. et al. SARS-CoV-2 triggers DNA damage response in Vero E6 cells. Biochem. Biophys. Res. Commun. 579, 141–145 (2021).",
"volume": "579",
"year": "2021"
},
{
"DOI": "10.1165/rcmb.2021-0205le",
"doi-asserted-by": "publisher",
"key": "1096_CR9",
"unstructured": "Lipskaia, L. et al. Evidence that SARS-CoV-2 induces lung-cell senescence: potential impact on COVID-19 lung disease. Am. J. Respir. Cell Mol. Biol. https://doi.org/10.1165/rcmb.2021-0205le (2021)."
},
{
"DOI": "10.1126/scitranslmed.abj7790",
"author": "F D’Agnillo",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Sci. Transl. Med.",
"key": "1096_CR10",
"unstructured": "D’Agnillo, F. et al. Lung epithelial and endothelial damage, loss of tissue repair, inhibition of fibrinolysis, and cellular senescence in fatal COVID-19. Sci. Transl. Med. 13, 1–18 (2021).",
"volume": "13",
"year": "2021"
},
{
"DOI": "10.1038/nature08467",
"author": "SP Jackson",
"doi-asserted-by": "publisher",
"first-page": "1071",
"journal-title": "Nature",
"key": "1096_CR11",
"unstructured": "Jackson, S. P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).",
"volume": "461",
"year": "2009"
},
{
"DOI": "10.1101/gad.2021311",
"author": "SE Polo",
"doi-asserted-by": "publisher",
"first-page": "409",
"journal-title": "Genes Dev.",
"key": "1096_CR12",
"unstructured": "Polo, S. E. & Jackson, S. P. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 25, 409–433 (2011).",
"volume": "25",
"year": "2011"
},
{
"DOI": "10.1038/nrc2440",
"author": "F d’Adda di Fagagna",
"doi-asserted-by": "publisher",
"first-page": "512",
"journal-title": "Nat. Rev. Cancer",
"key": "1096_CR13",
"unstructured": "d’Adda di Fagagna, F. Living on a break: cellular senescence as a DNA-damage response. Nat. Rev. Cancer 8, 512–522 (2008).",
"volume": "8",
"year": "2008"
},
{
"DOI": "10.1038/s41580-020-00314-w",
"author": "R Di Micco",
"doi-asserted-by": "publisher",
"first-page": "75",
"journal-title": "Nat. Rev. Mol. Cell Biol.",
"key": "1096_CR14",
"unstructured": "Di Micco, R., Krizhanovsky, V., Baker, D. & d’Adda di Fagagna, F. Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 22, 75–95 (2021).",
"volume": "22",
"year": "2021"
},
{
"DOI": "10.1038/ncb1909",
"author": "F Rodier",
"doi-asserted-by": "publisher",
"first-page": "973",
"journal-title": "Nat. Cell Biol.",
"key": "1096_CR15",
"unstructured": "Rodier, F. et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 11, 973–979 (2009).",
"volume": "11",
"year": "2009"
},
{
"DOI": "10.1038/ncb3643",
"author": "F Michelini",
"doi-asserted-by": "publisher",
"first-page": "1400",
"journal-title": "Nat. Cell Biol.",
"key": "1096_CR16",
"unstructured": "Michelini, F. et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 19, 1400–1411 (2017).",
"volume": "19",
"year": "2017"
},
{
"DOI": "10.1038/s41556-019-0392-4",
"author": "F Pessina",
"doi-asserted-by": "publisher",
"first-page": "1286",
"journal-title": "Nat. Cell Biol.",
"key": "1096_CR17",
"unstructured": "Pessina, F. et al. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat. Cell Biol. 21, 1286–1299 (2019).",
"volume": "21",
"year": "2019"
},
{
"DOI": "10.1038/nature11179",
"doi-asserted-by": "publisher",
"key": "1096_CR18",
"unstructured": "Francia, S. et al. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature https://doi.org/10.1038/nature11179 (2012)."
},
{
"DOI": "10.15252/embj.2018101379",
"doi-asserted-by": "publisher",
"key": "1096_CR19",
"unstructured": "Kilic, S. et al. Phase separation of 53 BP 1 determines liquid-like behavior of DNA repair compartments. EMBO J. https://doi.org/10.15252/embj.2018101379 (2019)."
},
{
"DOI": "10.1038/s41467-018-07799-2",
"author": "G D’Alessandro",
"doi-asserted-by": "publisher",
"journal-title": "Nat. Commun.",
"key": "1096_CR20",
"unstructured": "D’Alessandro, G. et al. BRCA2 controls DNA:RNA hybrid level at DSBs by mediating RNase H2 recruitment. Nat. Commun. 9, 5376 (2018).",
"volume": "9",
"year": "2018"
},
{
"DOI": "10.1101/2020.06.18.160648",
"doi-asserted-by": "publisher",
"key": "1096_CR21",
"unstructured": "Savastano, A., de Opakua, A. I., Rankovic, M. & Zweckstetter, M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat. Commun. https://doi.org/10.1101/2020.06.18.160648 (2020)."
},
{
"DOI": "10.15252/embj.2020106478",
"doi-asserted-by": "publisher",
"key": "1096_CR22",
"unstructured": "Perdikari, T. M. et al. SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J. https://doi.org/10.15252/embj.2020106478 (2020)."
},
{
"DOI": "10.1016/j.bbrc.2008.06.112",
"author": "H Naruyama",
"doi-asserted-by": "publisher",
"first-page": "79",
"journal-title": "Biochem. Biophys. Res. Commun.",
"key": "1096_CR23",
"unstructured": "Naruyama, H. et al. Essential role of Chk1 in S phase progression through regulation of RNR2 expression. Biochem. Biophys. Res. Commun. 374, 79–83 (2008).",
"volume": "374",
"year": "2008"
},
{
"DOI": "10.1038/s41577-022-00785-2",
"doi-asserted-by": "publisher",
"key": "1096_CR24",
"unstructured": "Schmitt, C. A. et al. COVID-19 and cellular senescence. Nat. Rev. Immunol. https://doi.org/10.1038/s41577-022-00785-2 (2022)."
},
{
"DOI": "10.1038/s41564-020-00846-z",
"doi-asserted-by": "publisher",
"key": "1096_CR25",
"unstructured": "Schmidt, N. et al. The SARS-CoV-2 RNA–protein interactome in infected human cells. Nat. Microbiol. https://doi.org/10.1038/s41564-020-00846-z (2020)."
},
{
"DOI": "10.1371/journal.pbio.3001490",
"author": "A Kratzel",
"doi-asserted-by": "publisher",
"first-page": "e300149",
"journal-title": "PLoS Biol.",
"key": "1096_CR26",
"unstructured": "Kratzel, A. et al. A genome-wide CRISPR screen identifies interactors of the autophagy pathway as conserved coronavirus targets. PLoS Biol. 19, e300149 (2021).",
"volume": "19",
"year": "2021"
},
{
"DOI": "10.1074/jbc.M303952200",
"author": "A Koç",
"doi-asserted-by": "publisher",
"first-page": "223",
"journal-title": "J. Biol. Chem.",
"key": "1096_CR27",
"unstructured": "Koç, A., Wheeler, L. J., Mathews, C. K. & Merrill, G. F. Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J. Biol. Chem. 279, 223–230 (2004).",
"volume": "279",
"year": "2004"
},
{
"DOI": "10.1016/j.dnarep.2004.03.020",
"author": "D Shechter",
"doi-asserted-by": "publisher",
"first-page": "901",
"journal-title": "DNA Repair",
"key": "1096_CR28",
"unstructured": "Shechter, D., Costanzo, V. & Gautier, J. Regulation of DNA replication by ATR: signaling in response to DNA intermediates. DNA Repair 3, 901–908 (2004).",
"volume": "3",
"year": "2004"
},
{
"DOI": "10.26508/lsa.202000786",
"author": "D Bestle",
"doi-asserted-by": "publisher",
"first-page": "e202000786",
"journal-title": "Life Sci. Alliance",
"key": "1096_CR29",
"unstructured": "Bestle, D. et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance 3, e202000786 (2020).",
"volume": "3",
"year": "2020"
},
{
"DOI": "10.1073/pnas.2022643118",
"author": "Y Li",
"doi-asserted-by": "publisher",
"first-page": "e2022643118",
"journal-title": "Proc. Natl Acad. Sci. USA",
"key": "1096_CR30",
"unstructured": "Li, Y. et al. SARS-CoV-2 induces double-stranded RNA-mediated innate immune responses in respiratory epithelial-derived cells and cardiomyocytes. Proc. Natl Acad. Sci. USA 118, e2022643118 (2021).",
"volume": "118",
"year": "2021"
},
{
"DOI": "10.1038/nature23470",
"author": "SM Harding",
"doi-asserted-by": "publisher",
"first-page": "466",
"journal-title": "Nature",
"key": "1096_CR31",
"unstructured": "Harding, S. M. et al. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 (2017).",
"volume": "548",
"year": "2017"
},
{
"DOI": "10.1002/hep.28685",
"author": "MK Thomsen",
"doi-asserted-by": "publisher",
"first-page": "746",
"journal-title": "Hepatology",
"key": "1096_CR32",
"unstructured": "Thomsen, M. K. et al. Lack of immunological DNA sensing in hepatocytes facilitates hepatitis B virus infection. Hepatology 64, 746–759 (2016).",
"volume": "64",
"year": "2016"
},
{
"DOI": "10.1038/s41586-021-03995-1",
"author": "S Lee",
"doi-asserted-by": "publisher",
"first-page": "283",
"journal-title": "Nature",
"key": "1096_CR33",
"unstructured": "Lee, S. et al. Virus-induced senescence is driver and therapeutic target in COVID-19. Nature 599, 283–289 (2021).",
"volume": "599",
"year": "2021"
},
{
"DOI": "10.1183/13993003.02951-2021",
"doi-asserted-by": "publisher",
"key": "1096_CR34",
"unstructured": "Evangelou, K. et al. Pulmonary infection by SARS-CoV-2 induces senescence accompanied by an inflammatory phenotype in severe COVID-19: possible implications for viral mutagenesis. Eur. Respir. J. https://doi.org/10.1183/13993003.02951-2021 (2022)."
},
{
"DOI": "10.15252/embj.2018101284",
"author": "MA González Besteiro",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "EMBO J.",
"key": "1096_CR35",
"unstructured": "González Besteiro, M. A. et al. Chk1 loss creates replication barriers that compromise cell survival independently of excess origin firing. EMBO J. 38, 1–16 (2019).",
"volume": "38",
"year": "2019"
},
{
"DOI": "10.1074/jbc.M109.003020",
"author": "YW Zhang",
"doi-asserted-by": "publisher",
"first-page": "18085",
"journal-title": "J. Biol. Chem.",
"key": "1096_CR36",
"unstructured": "Zhang, Y. W., Jones, T. L., Martin, S. E., Caplen, N. J. & Pommier, Y. Implication of checkpoint kinase-dependent up-regulation of ribonucleotide reductase R2 in DNA damage response. J. Biol. Chem. 284, 18085–18095 (2009).",
"volume": "284",
"year": "2009"
},
{
"DOI": "10.1101/gad.1482106",
"author": "CL Sansam",
"doi-asserted-by": "publisher",
"first-page": "3117",
"journal-title": "Genes Dev.",
"key": "1096_CR37",
"unstructured": "Sansam, C. L. et al. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev. 20, 3117–3129 (2006).",
"volume": "20",
"year": "2006"
},
{
"DOI": "10.1038/ncb1061",
"author": "LAA Higa",
"doi-asserted-by": "publisher",
"first-page": "1008",
"journal-title": "Nat. Cell Biol.",
"key": "1096_CR38",
"unstructured": "Higa, L. A. A., Mihaylov, I. S., Banks, D. P., Zheng, J. & Zhang, H. Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 5, 1008–1015 (2003).",
"volume": "5",
"year": "2003"
},
{
"DOI": "10.1038/ncb1172",
"author": "J Hu",
"doi-asserted-by": "publisher",
"first-page": "1003",
"journal-title": "Nat. Cell Biol.",
"key": "1096_CR39",
"unstructured": "Hu, J., McCall, C. M., Ohta, T. & Xiong, Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 6, 1003–1009 (2004).",
"volume": "6",
"year": "2004"
},
{
"DOI": "10.1038/s41586-020-2286-9",
"author": "DE Gordon",
"doi-asserted-by": "publisher",
"first-page": "459",
"journal-title": "Nature",
"key": "1096_CR40",
"unstructured": "Gordon, D. E. et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468 (2020).",
"volume": "583",
"year": "2020"
},
{
"DOI": "10.1073/pnas.2016650117",
"author": "L Miorin",
"doi-asserted-by": "publisher",
"first-page": "28344",
"journal-title": "Proc. Natl Acad. Sci. USA",
"key": "1096_CR41",
"unstructured": "Miorin, L. et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl Acad. Sci. USA 117, 28344–28354 (2020).",
"volume": "117",
"year": "2020"
},
{
"DOI": "10.1128/mBio.00065-21",
"author": "A Addetia",
"doi-asserted-by": "publisher",
"first-page": "e00065-21",
"journal-title": "mBio",
"key": "1096_CR42",
"unstructured": "Addetia, A. et al. SARS-CoV-2 orf6 disrupts bidirectional nucleocytoplasmic transport through interactions with rae1 and nup98. mBio 12, e00065-21 (2021).",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1074/jbc.M112.350397",
"author": "J Wang",
"doi-asserted-by": "publisher",
"first-page": "25501",
"journal-title": "J. Biol. Chem.",
"key": "1096_CR43",
"unstructured": "Wang, J., Han, X., Feng, X., Wang, Z. & Zhang, Y. Coupling cellular localization and function of checkpoint kinase 1 (Chk1) in checkpoints and cell viability. J. Biol. Chem. 287, 25501–25509 (2012).",
"volume": "287",
"year": "2012"
},
{
"DOI": "10.4049/jimmunol.2100684",
"author": "C Sui",
"doi-asserted-by": "publisher",
"first-page": "753",
"journal-title": "J. Immunol.",
"key": "1096_CR44",
"unstructured": "Sui, C. et al. SARS-CoV-2 NSP13 inhibits type I IFN production by degradation of TBK1 via p62-dependent selective autophagy. J. Immunol. 208, 753–761 (2022).",
"volume": "208",
"year": "2022"
},
{
"DOI": "10.1080/15548627.2020.1797280",
"author": "DJ Klionsky",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Autophagy",
"key": "1096_CR45",
"unstructured": "Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 17, 1–382 (2021).",
"volume": "17",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2014.02.049",
"author": "DR Green",
"doi-asserted-by": "publisher",
"first-page": "65",
"journal-title": "Cell",
"key": "1096_CR46",
"unstructured": "Green, D. R. & Levine, B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell 157, 65–75 (2014).",
"volume": "157",
"year": "2014"
},
{
"DOI": "10.1038/s41422-020-00408-2",
"doi-asserted-by": "publisher",
"key": "1096_CR47",
"unstructured": "Chen, H. et al. Liquid–liquid phase separation by SARS-CoV-2 nucleocapsid protein and RNA. Cell Res. https://doi.org/10.1038/s41422-020-00408-2 (2020)."
},
{
"DOI": "10.1038/s41421-020-00240-3",
"author": "J Wang",
"doi-asserted-by": "publisher",
"first-page": "3",
"journal-title": "Cell Discov.",
"key": "1096_CR48",
"unstructured": "Wang, J., Shi, C., Xu, Q. & Yin, H. SARS-CoV-2 nucleocapsid protein undergoes liquid–liquid phase separation into stress granules through its N-terminal intrinsically disordered region. Cell Discov. 7, 3–7 (2021).",
"volume": "7",
"year": "2021"
},
{
"DOI": "10.1038/s41556-021-00710-0",
"author": "S Wang",
"doi-asserted-by": "publisher",
"first-page": "718",
"journal-title": "Nat. Cell Biol.",
"key": "1096_CR49",
"unstructured": "Wang, S. et al. Targeting liquid–liquid phase separation of SARS-CoV-2 nucleocapsid protein promotes innate antiviral immunity by elevating MAVS activity. Nat. Cell Biol. 23, 718–732 (2021).",
"volume": "23",
"year": "2021"
},
{
"DOI": "10.1083/jcb.200503043",
"author": "S Bekker-Jensen",
"doi-asserted-by": "publisher",
"first-page": "201",
"journal-title": "J. Cell Biol.",
"key": "1096_CR50",
"unstructured": "Bekker-Jensen, S., Lukas, C., Melander, F., Bartek, J. & Lukas, J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J. Cell Biol. 170, 201–211 (2005).",
"volume": "170",
"year": "2005"
},
{
"DOI": "10.1038/s43587-022-00170-7",
"author": "S Tsuji",
"doi-asserted-by": "publisher",
"first-page": "115",
"journal-title": "Nat. Aging",
"key": "1096_CR51",
"unstructured": "Tsuji, S. et al. SARS-CoV-2 infection triggers paracrine senescence and leads to a sustained senescence-associated inflammatory response. Nat. Aging 2, 115–124 (2022).",
"volume": "2",
"year": "2022"
},
{
"DOI": "10.15252/msb.202110396",
"author": "X Liu",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Mol. Syst. Biol.",
"key": "1096_CR52",
"unstructured": "Liu, X. et al. SARS‐CoV‐2–host proteome interactions for antiviral drug discovery. Mol. Syst. Biol. 17, 1–26 (2021).",
"volume": "17",
"year": "2021"
},
{
"DOI": "10.1371/journal.pbio.3001158",
"author": "P V’kovski",
"doi-asserted-by": "publisher",
"first-page": "e3001158",
"journal-title": "PLoS Biol.",
"key": "1096_CR53",
"unstructured": "V’kovski, P. et al. Disparate temperature-dependent virus–host dynamics for SARS-CoV-2 and SARS-CoV in the human respiratory epithelium. PLoS Biol. 19, e3001158 (2021).",
"volume": "19",
"year": "2021"
},
{
"DOI": "10.1016/j.tcb.2013.09.003",
"author": "M Zimmermann",
"doi-asserted-by": "publisher",
"first-page": "108",
"journal-title": "Trends Cell Biol.",
"key": "1096_CR54",
"unstructured": "Zimmermann, M. & De Lange, T. 53BP1: pro choice in DNA repair. Trends Cell Biol. 24, 108–117 (2014).",
"volume": "24",
"year": "2014"
},
{
"author": "A Gunn",
"first-page": "588",
"journal-title": "Methods Mol. Biol.",
"key": "1096_CR55",
"unstructured": "Gunn, A. & Stark, J. M. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol. Biol. 531, 588 (2012).",
"volume": "531",
"year": "2012"
},
{
"DOI": "10.1038/s41598-019-42892-6",
"author": "U Gioia",
"doi-asserted-by": "publisher",
"journal-title": "Sci. Rep.",
"key": "1096_CR56",
"unstructured": "Gioia, U. et al. Pharmacological boost of DNA damage response and repair by enhanced biogenesis of DNA damage response RNAs. Sci. Rep. 9, 6460 (2019).",
"volume": "9",
"year": "2019"
},
{
"DOI": "10.1038/ncomms13785",
"author": "RA Shamanna",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Nat. Commun.",
"key": "1096_CR57",
"unstructured": "Shamanna, R. A. et al. WRN regulates pathway choice between classical and alternative non-homologous end joining. Nat. Commun. 7, 1–12 (2016).",
"volume": "7",
"year": "2016"
},
{
"DOI": "10.1016/j.ebiom.2020.103104",
"author": "R Bussani",
"doi-asserted-by": "publisher",
"first-page": "103104",
"journal-title": "EBioMedicine",
"key": "1096_CR58",
"unstructured": "Bussani, R. et al. Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. EBioMedicine 61, 103104 (2020).",
"volume": "61",
"year": "2020"
},
{
"DOI": "10.1146/annurev.micro.112408.134016",
"author": "MD Weitzman",
"doi-asserted-by": "publisher",
"first-page": "61",
"journal-title": "Annu. Rev. Microbiol.",
"key": "1096_CR59",
"unstructured": "Weitzman, M. D., Lilley, C. E. & Chaurushiya, M. S. Genomes in conflict: maintaining genome integrity during virus infection. Annu. Rev. Microbiol. 64, 61–81 (2010).",
"volume": "64",
"year": "2010"
},
{
"DOI": "10.1016/j.cell.2020.10.004",
"author": "AK Banerjee",
"doi-asserted-by": "publisher",
"first-page": "1325",
"journal-title": "Cell",
"key": "1096_CR60",
"unstructured": "Banerjee, A. K. et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell 183, 1325–1339.e21 (2020).",
"volume": "183",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.04.026",
"author": "D Blanco-Melo",
"doi-asserted-by": "publisher",
"first-page": "1036",
"journal-title": "Cell",
"key": "1096_CR61",
"unstructured": "Blanco-Melo, D. et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181, 1036–1045.e9 (2020).",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1186/s13062-021-00305-7",
"author": "H Ren",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Biol. Direct",
"key": "1096_CR62",
"unstructured": "Ren, H. et al. Micronucleus production, activation of DNA damage response and cGAS-STING signaling in syncytia induced by SARS-CoV-2 infection. Biol. Direct 16, 1–10 (2021).",
"volume": "16",
"year": "2021"
},
{
"DOI": "10.1038/s41392-021-00800-3",
"doi-asserted-by": "publisher",
"key": "1096_CR63",
"unstructured": "Zhou, Z. et al. Sensing of cytoplasmic chromatin by cGAS activates innate immune response in SARS-CoV-2 infection. Signal Transduct. Target. Ther. https://doi.org/10.1038/s41392-021-00800-3 (2021)."
},
{
"DOI": "10.3390/v13102056",
"doi-asserted-by": "publisher",
"key": "1096_CR64",
"unstructured": "Jiang, H. & Mei, Y. F. SARS-CoV -2 spike impairs DNA damage repair and inhibits V(D)J recombination in vitro. Viruses https://doi.org/10.3390/v13102056 (2021)."
},
{
"DOI": "10.1016/j.celrep.2021.108940",
"author": "G Garcia",
"doi-asserted-by": "publisher",
"first-page": "108940",
"journal-title": "Cell Rep.",
"key": "1096_CR65",
"unstructured": "Garcia, G. et al. Antiviral drug screen identifies DNA-damage response inhibitor as potent blocker of SARS-CoV-2 replication. Cell Rep. 35, 108940 (2021).",
"volume": "35",
"year": "2021"
},
{
"DOI": "10.1099/vir.0.044412-0",
"author": "AS Turnell",
"doi-asserted-by": "publisher",
"first-page": "2076",
"journal-title": "J. Gen. Virol.",
"key": "1096_CR66",
"unstructured": "Turnell, A. S. & Grand, R. J. DNA viruses and the cellular DNA-damage response. J. Gen. Virol. 93, 2076–2097 (2012).",
"volume": "93",
"year": "2012"
},
{
"DOI": "10.1073/pnas.1603435113",
"author": "Y Ma-Lauer",
"doi-asserted-by": "publisher",
"first-page": "E5192",
"journal-title": "Proc. Natl Acad. Sci. USA",
"key": "1096_CR67",
"unstructured": "Ma-Lauer, Y. et al. P53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1. Proc. Natl Acad. Sci. USA 113, E5192–E5201 (2016).",
"volume": "113",
"year": "2016"
},
{
"DOI": "10.1038/emboj.2009.400",
"author": "CE Lilley",
"doi-asserted-by": "publisher",
"first-page": "943",
"journal-title": "EMBO J.",
"key": "1096_CR68",
"unstructured": "Lilley, C. E. et al. A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses. EMBO J. 29, 943–955 (2010).",
"volume": "29",
"year": "2010"
},
{
"DOI": "10.1038/nrm714",
"author": "JM Trimarchi",
"doi-asserted-by": "publisher",
"first-page": "11",
"journal-title": "Nat. Rev. Mol. Cell Biol.",
"key": "1096_CR69",
"unstructured": "Trimarchi, J. M. & Lees, J. A. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3, 11–20 (2002).",
"volume": "3",
"year": "2002"
},
{
"DOI": "10.1016/j.bbagrm.2016.02.012",
"author": "C Gong",
"doi-asserted-by": "publisher",
"first-page": "612",
"journal-title": "Biochim. Biophys. Acta Gene Regul. Mech.",
"key": "1096_CR70",
"unstructured": "Gong, C. et al. ATR-CHK1-E2F3 signaling transactivates human ribonucleotide reductase small subunit M2 for DNA repair induced by the chemical carcinogen MNNG. Biochim. Biophys. Acta Gene Regul. Mech. 1859, 612–626 (2016).",
"volume": "1859",
"year": "2016"
},
{
"DOI": "10.1016/j.cell.2020.04.011",
"author": "D Kim",
"doi-asserted-by": "publisher",
"first-page": "914",
"journal-title": "Cell",
"key": "1096_CR71",
"unstructured": "Kim, D. et al. The architecture of SARS-CoV-2 transcriptome. Cell 181, 914–921.e10 (2020).",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1016/j.virol.2016.09.028",
"author": "DC Anacker",
"doi-asserted-by": "publisher",
"first-page": "383",
"journal-title": "Virology",
"key": "1096_CR72",
"unstructured": "Anacker, D. C. et al. HPV31 utilizes the ATR-Chk1 pathway to maintain elevated RRM2 levels and a replication-competent environment in differentiating keratinocytes. Virology 499, 383–396 (2016).",
"volume": "499",
"year": "2016"
},
{
"DOI": "10.1016/j.virusres.2005.05.007",
"author": "KA Timani",
"doi-asserted-by": "publisher",
"first-page": "23",
"journal-title": "Virus Res.",
"key": "1096_CR73",
"unstructured": "Timani, K. A. et al. Nuclear/nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus. Virus Res. 114, 23–34 (2005).",
"volume": "114",
"year": "2005"
},
{
"DOI": "10.1128/JVI.79.17.11507-11512.2005",
"author": "RRR Rowland",
"doi-asserted-by": "publisher",
"first-page": "11507",
"journal-title": "J. Virol.",
"key": "1096_CR74",
"unstructured": "Rowland, R. R. R. et al. Intracellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein: absence of nucleolar accumulation during infection and after expression as a recombinant protein in Vero cells. J. Virol. 79, 11507–11512 (2005).",
"volume": "79",
"year": "2005"
},
{
"DOI": "10.1073/pnas.2008176117",
"author": "AB Gussow",
"doi-asserted-by": "publisher",
"first-page": "15193",
"journal-title": "Proc. Natl Acad. Sci. USA",
"key": "1096_CR75",
"unstructured": "Gussow, A. B. et al. Genomic determinants of pathogenicity in SARS-CoV-2 and other human coronaviruses. Proc. Natl Acad. Sci. USA 117, 15193–15199 (2020).",
"volume": "117",
"year": "2020"
},
{
"DOI": "10.1038/s41598-021-89605-6",
"author": "A Ahmadi",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Sci. Rep.",
"key": "1096_CR76",
"unstructured": "Ahmadi, A. & Moradi, S. In silico analysis suggests the RNAi-enhancing antibiotic enoxacin as a potential inhibitor of SARS-CoV-2 infection. Sci. Rep. 11, 1–14 (2021).",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.1016/S0140-6736(20)30628-0",
"author": "P Mehta",
"doi-asserted-by": "publisher",
"first-page": "1033",
"journal-title": "Lancet",
"key": "1096_CR77",
"unstructured": "Mehta, P. et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 395, 1033–1034 (2020).",
"volume": "395",
"year": "2020"
},
{
"DOI": "10.1084/jem.20180139",
"author": "T Li",
"doi-asserted-by": "publisher",
"first-page": "1287",
"journal-title": "J. Exp. Med.",
"key": "1096_CR78",
"unstructured": "Li, T. & Chen, Z. J. The cGAS–cGAMP–STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 215, 1287–1299 (2018).",
"volume": "215",
"year": "2018"
},
{
"DOI": "10.1016/j.celrep.2013.03.004",
"author": "KM Aird",
"doi-asserted-by": "publisher",
"first-page": "1252",
"journal-title": "Cell Rep.",
"key": "1096_CR79",
"unstructured": "Aird, K. M. et al. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Rep. 3, 1252–1265 (2013).",
"volume": "3",
"year": "2013"
},
{
"DOI": "10.1038/s43587-022-00321-w",
"author": "M Mavrikaki",
"doi-asserted-by": "publisher",
"first-page": "1130",
"journal-title": "Nat. Aging",
"key": "1096_CR80",
"unstructured": "Mavrikaki, M., Lee, J. D., Solomon, I. H. & Slack, F. J. Severe COVID-19 is associated with molecular signatures of aging in the human brain. Nat. Aging 2, 1130–1137 (2022).",
"volume": "2",
"year": "2022"
},
{
"author": "X Cao",
"first-page": "1",
"journal-title": "Nat. Commun.",
"key": "1096_CR81",
"unstructured": "Cao, X. et al. Accelerated biological aging in COVID-19 patients. Nat. Commun. 13, 1–7 (2022).",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1038/ncb2466",
"author": "M Fumagalli",
"doi-asserted-by": "publisher",
"first-page": "355",
"journal-title": "Nat. Cell Biol.",
"key": "1096_CR82",
"unstructured": "Fumagalli, M. et al. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat. Cell Biol. 14, 355–365 (2012).",
"volume": "14",
"year": "2012"
},
{
"DOI": "10.1016/j.gde.2014.06.009",
"author": "F Rossiello",
"doi-asserted-by": "publisher",
"first-page": "89",
"journal-title": "Curr. Opin. Genet. Dev.",
"key": "1096_CR83",
"unstructured": "Rossiello, F., Herbig, U., Longhese, M. P. & Fumagalli, M. & d’Adda di Fagagna, F. Irreparable telomeric DNA damage and persistent DDR signalling as a shared causative mechanism of cellular senescence and ageing. Curr. Opin. Genet. Dev. 26, 89–95 (2014).",
"volume": "26",
"year": "2014"
},
{
"DOI": "10.1038/s41591-021-01283-z",
"author": "A Nalbandian",
"doi-asserted-by": "publisher",
"first-page": "601",
"journal-title": "Nat. Med.",
"key": "1096_CR84",
"unstructured": "Nalbandian, A. et al. Post-acute COVID-19 syndrome. Nat. Med. 27, 601–615 (2021).",
"volume": "27",
"year": "2021"
},
{
"DOI": "10.1126/sciimmunol.abl9929",
"author": "V Fumagalli",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Sci. Immunol.",
"key": "1096_CR85",
"unstructured": "Fumagalli, V. et al. Administration of aerosolized SARS-CoV-2 to K18-hACE2 mice uncouples respiratory infection from fatal neuroinvasion. Sci. Immunol. 7, 1–11 (2022).",
"volume": "7",
"year": "2022"
},
{
"DOI": "10.1128/JVI.00543-20",
"author": "D Licastro",
"doi-asserted-by": "publisher",
"first-page": "9",
"journal-title": "J. Virol.",
"key": "1096_CR86",
"unstructured": "Licastro, D. et al. Isolation and full-length genome characterization of SARS-CoV- 2 from COVID-19 cases in Northern Italy. J. Virol. 94, 9–12 (2020).",
"volume": "94",
"year": "2020"
},
{
"DOI": "10.1073/pnas.92.20.9363",
"author": "GP Dimri",
"doi-asserted-by": "publisher",
"first-page": "9363",
"journal-title": "Proc. Natl Acad. Sci. USA",
"key": "1096_CR87",
"unstructured": "Dimri, G. P. et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA 92, 9363–9367 (1995).",
"volume": "92",
"year": "1995"
},
{
"DOI": "10.1074/jbc.M112.441147",
"author": "M Frangini",
"doi-asserted-by": "publisher",
"first-page": "5624",
"journal-title": "J. Biol. Chem.",
"key": "1096_CR88",
"unstructured": "Frangini, M. et al. Synthesis of mitochondrial DNA precursors during myogenesis, an analysis in purified C2C12 myotubes. J. Biol. Chem. 288, 5624–5635 (2013).",
"volume": "288",
"year": "2013"
},
{
"DOI": "10.1093/nar/gkp1141",
"author": "P Ferraro",
"doi-asserted-by": "publisher",
"first-page": "e85",
"journal-title": "Nucleic Acids Res.",
"key": "1096_CR89",
"unstructured": "Ferraro, P., Franzolin, E., Pontarin, G., Reichard, P. & Bianchi, V. Quantitation of cellular deoxynucleoside triphosphates. Nucleic Acids Res. 38, e85–e85 (2010).",
"volume": "38",
"year": "2010"
},
{
"DOI": "10.1073/pnas.1211289109",
"author": "G Pontarin",
"doi-asserted-by": "publisher",
"first-page": "13302",
"journal-title": "Proc. Natl Acad. Sci. USA",
"key": "1096_CR90",
"unstructured": "Pontarin, G., Ferraro, P., Bee, L., Reichard, P. & Bianchi, V. Mammalian ribonucleotide reductase subunit p53R2 is required for mitochondrial DNA replication and DNA repair in quiescent cells. Proc. Natl Acad. Sci. USA 109, 13302–13307 (2012).",
"volume": "109",
"year": "2012"
},
{
"DOI": "10.1158/2326-6066.CIR-20-0645",
"author": "G Morello",
"doi-asserted-by": "publisher",
"first-page": "825",
"journal-title": "Cancer Immunol. Res.",
"key": "1096_CR91",
"unstructured": "Morello, G. et al. T cells expressing receptor recombination/revision machinery are detected in the tumor microenvironment and expanded in genomically over-unstable models. Cancer Immunol. Res. 9, 825–837 (2021).",
"volume": "9",
"year": "2021"
},
{
"DOI": "10.15252/embr.202153658",
"doi-asserted-by": "publisher",
"key": "1096_CR92",
"unstructured": "Sepe, S. et al. DNA damage response at telomeres boosts the transcription of SARS-CoV-2 receptor ACE2 during aging. EMBO Rep. https://doi.org/10.15252/embr.202153658 (2021)."
}
],
"reference-count": 92,
"references-count": 92,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.nature.com/articles/s41556-023-01096-x"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "SARS-CoV-2 infection induces DNA damage, through CHK1 degradation and impaired 53BP1 recruitment, and cellular senescence",
"type": "journal-article",
"update-policy": "https://doi.org/10.1007/springer_crossmark_policy",
"volume": "25"
}