The SARS-CoV-2 main protease causes mitochondrial dysfunction in a yeast model
et al., Scientific Reports, doi:10.1038/s41598-025-11993-w, Jul 2025
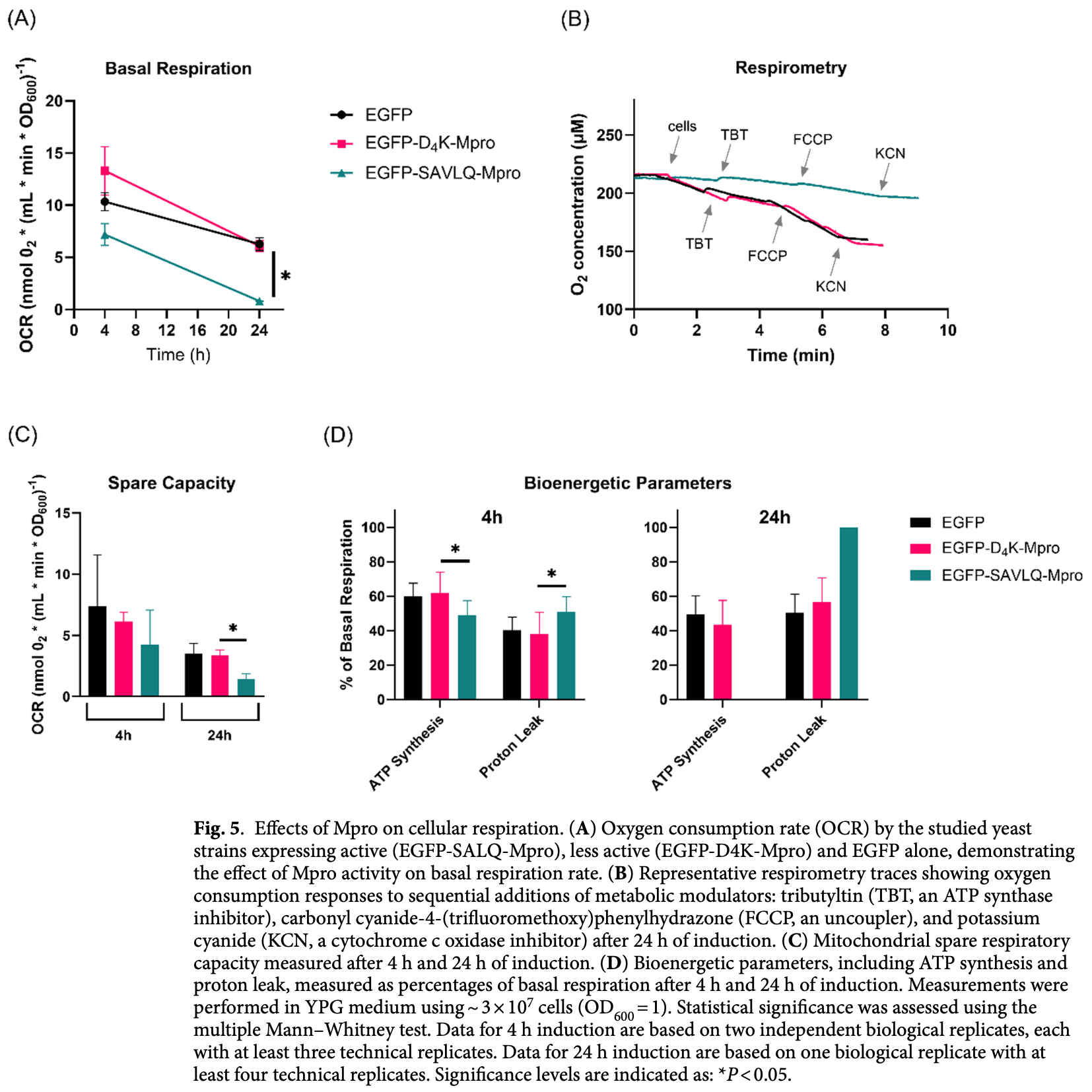
In vitro yeast study showing mitochondrial harm with SARS-CoV-2 main protease (Mpro). Authors expressed wild-type Mpro in Saccharomyces cerevisiae under a galactose-inducible promoter and found that active, self-cleaved Mpro was highly toxic: it halted growth on non-fermentable carbon sources and cut basal respiration and spare respiratory capacity by more than half, while confocal microscopy revealed early hyper-polarisation followed by persistent loss of mitochondrial membrane potential and fragmented organelles.
Grabiński et al., 18 Jul 2025, peer-reviewed, 5 authors.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
The SARS-CoV-2 main protease causes mitochondrial dysfunction in a yeast model
doi:10.1038/s41598-025-11993-w
Saccharomyces cerevisiae has proven to be an invaluable model organism for studying mitochondrial function owing to its genetic tractability and the high conservation of mitochondrial processes among eukaryotes, including humans. Yeasts are easy to culture and manipulate genetically, which allows rapid generation of mutant strains and detailed dissection of mitochondrial pathways. In addition, the ability of yeasts to survive without functional mitochondria allows the study of mutations that are lethal to organisms that are dependent on aerobic metabolism. Taking advantage of these benefits, we investigated the toxicity of SARS-CoV-2 main protease (Mpro) expression in yeast under conditions that enforce mitochondria-dependent aerobic metabolism. Our results showed that Mpro expression was highly toxic and significantly impaired yeast growth. Pronounced changes in the morphology and mitochondrial function were observed, indicating that mitochondrial pathways are exceptionally sensitive to Mpro activity. These results provide insights that may be relevant for understanding the effects of Mpro in more complex eukaryotic systems. Increasing amounts of data suggest that in cells infected with Betacoronavirus pandemicum (SARS-CoV-2), mitochondria are central to understanding COVID-19 pathogenesis. In human and rodent cells multiple mitochondrial changes, both structural and functional, have been found to have far-reaching consequences. These include hijacked mitochondria, reprogrammed metabolism, decreased oxidative phosphorylation (OXPHOS), increased reactive oxygen species (ROS) production, elevated mitochondrial iron levels, loss of mitochondrial integrity, cell death, disturbed autophagy, a reduced mitochondrial antiviral response (MAVS) and a type 1 interferon (IFN) response (see [1] [2] [3] ). Twenty out of 27 viral proteins have been implicated in these processes and have been shown to interact with host mitochondria 4, 5 . The SARS-CoV-2 main protease (Mpro), also known as 3-chymotrypsin-like protease (3CLpro), is a dimeric cysteine protease that is responsible for the cleavage of the viral polyproteins pp1a and pp1b, which are required for viral replication and transcription 6 . This enzyme shows no homology to human proteases and is thus an interesting drug target. As viral proteases cleave host proteins to hinder host immune responses and promote viral replication, numerous host proteins are predicted, via both computational and experimental methods, to be targeted by Mpro 7 . The human proteins confirmed to be cleaved include RNA polymerase II-associated protein 1 (RPAP1), Interleukin-1 receptor-associated kinase 1 (IRAK-1), nucleotide-binding oligomerization domain-like receptor (NLR), Solute carrier family 25 member 22 (SLC25A22) 7 . Among other processes, Mpro activity has been shown to influence human cell transcription and translation, apoptosis, the DNA damage response, lipid metabolism, vesicle trafficking, and the innate immune..
Author contributions W.G. designed the study, constructed expression systems, and performed experiments. A.Ki. contributed to experimental work, conceptual development, and manuscript writing. K.F. performed experiments. T.S. developed methodology and conducted data visualization. A.Ka. acquired funding, supervised the project, contributed to the conceptual framework, and co-wrote the manuscript.
Declarations Competing interests The authors declare no competing interests.
References
Alalam, A genetic trap in yeast for inhibitors of SARS-CoV-2 main protease, mSystems
Altmann, Westermann, Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae, Mol. Biol. Cell
Cao, Evaluation of SARS-CoV-2 main protease inhibitors using a novel Cell-Based assay, ACS Cent. Sci
Dejean, Beauvoit, Guérin, Rigoulet, Growth of the yeast Saccharomyces cerevisiae on a non-fermentable substrate: control of energetic yield by the amount of mitochondria, Biochim. Biophys. Acta
Escalante-Chong, Galactose metabolic genes in yeast respond to a ratio of galactose and glucose, Proc. Natl. Acad. Sci. U S A
Flynn, Comprehensive fitness landscape of SARS-CoV-2 Mpro reveals insights into viral resistance mechanisms, Elife
Gancedo, Gancedo, Sols, Glycerol metabolism in yeasts. Pathways of utilization and production, Eur. J. Biochem
Gałgańska, Viability of Saccharomyces cerevisiae cells following exposure to H2O2 and protective effect of Minocycline depend on the presence of VDAC, Eur. J. Pharmacol
Gordon, A SARS-CoV-2 protein interaction map reveals targets for drug repurposing, Nature
Grabiński, Meisoindigo: an effective inhibitor of SARS-CoV-2 main protease revealed by yeast system, BioRxiv, doi:10.1101/2023.09.03.555867
Guarnieri, Core mitochondrial genes are down-regulated during SARS-CoV-2 infection of rodent and human hosts, Sci. Transl Med
Guarnieri, SARS-CoV-2 mitochondrial metabolic and epigenomic reprogramming in COVID-19, Pharmacol. Res
Hughes, Hughes, Henderson, Yazvenko, Gottschling, Selective sorting and destruction of mitochondrial membrane proteins in aged yeast, eLife
Karachitos, Human VDAC isoforms differ in their capability to interact with Minocycline and to contribute to its cytoprotective activity, Mitochondrion
Klein, Swinnen, Thevelein, Nevoigt, Glycerol metabolism and transport in yeast and fungi: established knowledge and ambiguities, Environ. Microbiol
Lamphier, Ptashne, Multiple mechanisms mediate glucose repression of the yeast GAL1 gene, Proc. Natl. Acad. Sci. U S A
Laughery, Wyrick, Simple CRISPR-Cas9 genome editing in Saccharomyces cerevisiae, Curr. Protoc. Mol. Biol
Luo, Identification of human host substrates of the SARS-CoV-2 Mpro and PLpro using subtiligase N-Terminomics, ACS Infect. Dis
Melano, Lo, Su, Characterization of host substrates of SARS-CoV-2 main protease, Front. Microbiol
Merico, Ragni, Galafassi, Popolo, Compagno, Generation of an evolved Saccharomyces cerevisiae strain with a high freeze tolerance and an improved ability to grow on glycerol, J. Ind. Microbiol. Biotechnol
Mirdita, ColabFold: making protein folding accessible to all, Nat. Methods
Molnar, Mitochondrial dysfunction in long COVID: mechanisms, consequences, and potential therapeutic approaches, GeroScience
Ou, A yeast-based system to study SARS-CoV-2 Mpro structure and to identify nirmatrelvir resistant mutations, PLoS Pathog
Parmar, Structural differences in 3 C-like protease (Mpro) from SARS-CoV and SARS-CoV-2: molecular insights revealed by Molecular Dynamics Simulations, Struct. Chem, doi:10.1007/s11224-022-02089-6
Prasun, COVID-19: A mitochondrial perspective, DNA Cell. Biol
Randez-Gil, Sanz, Entian, Prieto, Carbon source-dependent phosphorylation of hexokinase PII and its role in the glucose-signaling response in yeast, Mol. Cell. Biol
Scott, Lacasse, Blom, Tonner, Blom, Predicted coronavirus Nsp5 protease cleavage sites in the human proteome, BMC Genomic Data
Singh, Chaubey, Chen, Suravajhala, Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis, Am. J. Physiol. Cell. Physiol
Stukalov, Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV, Nature
Swinnen, Re-evaluation of glycerol utilization in Saccharomyces cerevisiae: characterization of an isolate that grows on glycerol without supporting supplements, Biotechnol. Biofuels
Szögi, Novel biomarkers of mitochondrial dysfunction in long COVID patients, Geroscience h t t, doi:10.1007/s11357-024-01398-4
Ullrich, Nitsche, The SARS-CoV-2 main protease as drug target, Bioorg. Med. Chem. Lett
Xue, Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction, J. Mol. Biol
Yaghi, Andrews, Wylie, Iverson, High-Resolution substrate specificity profiling of SARS-CoV-2 mpro; comparison to SARS-CoV Mpro, ACS Chem. Biol