FGFR signaling and neddylation facilitate SARS-CoV-2 infection by modulating interferon induction and viral entry, respectively
et al., iScience, doi:10.1016/j.isci.2025.114566, Dec 2025
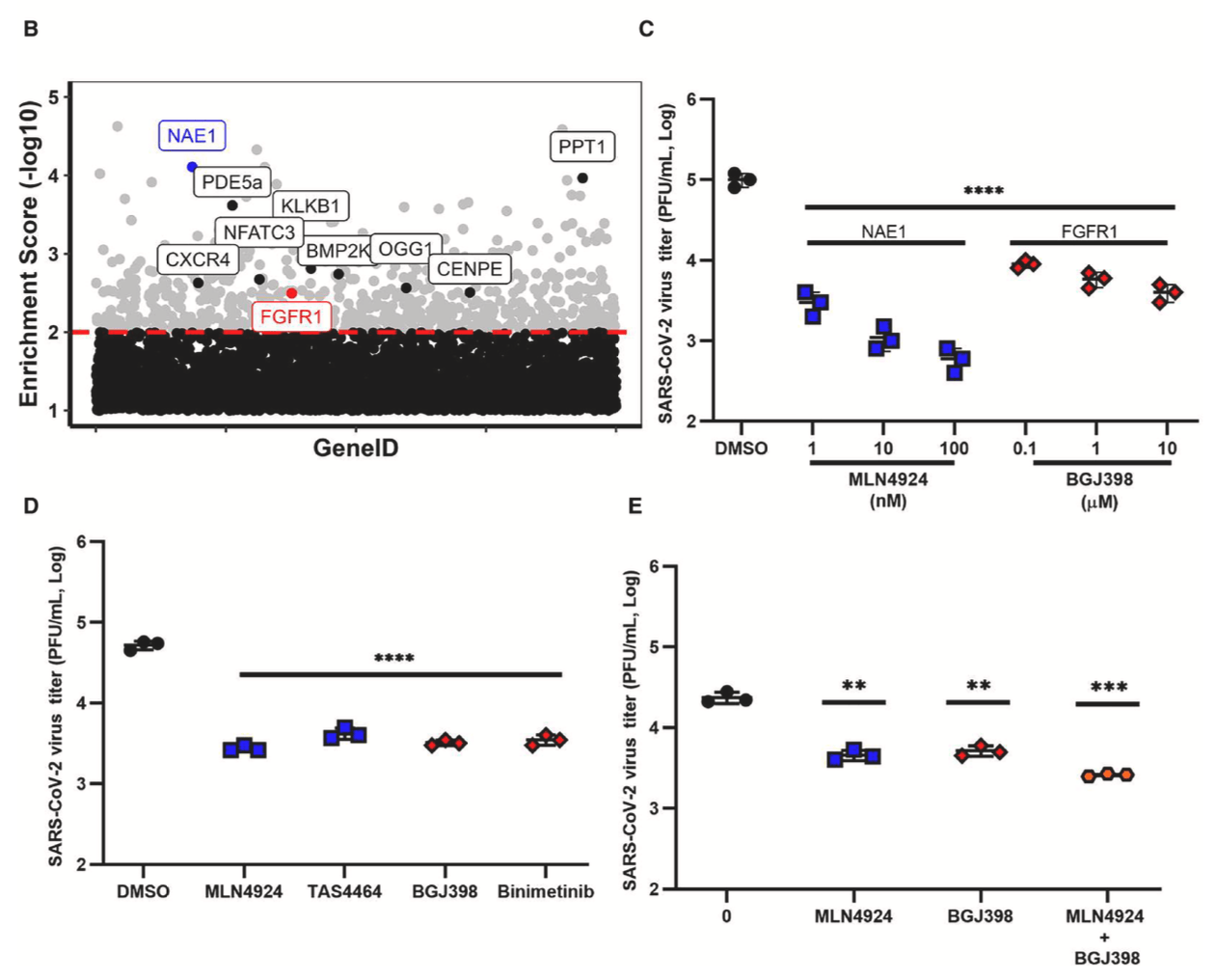
In vitro and mouse study showing that inhibitors of neddylation (MLN4924, TAS4464) and FGFR signaling (BGJ398, binimetinib) significantly reduce SARS-CoV-2 infection by targeting host dependency factors.
Felix-Lopez et al., 29 Dec 2025, Canada, peer-reviewed, 16 authors.
Contact: lopezoro@ualberta.ca (corresponding author), lopezoro@ualberta.ca (corresponding author), anil.kumar@usask.ca, thobman@ualberta.ca.
FGFR signaling and neddylation facilitate SARS-CoV-2 infection by modulating interferon induction and viral entry, respectively
iScience, doi:10.1016/j.isci.2025.114566
SARS-CoV-2 is the causative agent of COVID-19, and although vaccines have reduced disease severity, emerging variants remain a significant public health issue. Broadly effective therapeutics, particularly those targeting host pathways essential for coronavirus infection, are still needed. Here, we used a CRISPR knockout screen to identify druggable host factors required for SARS-CoV-2 infection. The screen revealed NAE1 and FGFR1 as key contributors to infection. Inhibitors, either FDA-approved or those in clinical trials, of these pathways reduced replication of both ancestral and contemporary viral variants. Mechanistic studies showed that FGFR1 promotes viral replication through downstream MEK/ERK signaling, while neddylation appears to support viral entry or infectivity rather than replication itself. In a murine model of severe COVID-19, inhibitors of NAE1 and FGFR1 significantly decreased viral load and lung pathology. These findings support the development of host-targeted antiviral strategies.
AUTHOR CONTRIBUTIONS A.F.-L., J.L.-O., A.K., and T.C.H. designed and managed the overall project. A.F.-L. drafted the original manuscript. A.F.-L., J.L.-O., and N.F. performed and analyzed in vitro experiments. A.F.-L., M.E., and Z.X. performed and analyzed in vivo experiments. B.B.H. analyzed the histopathology data. R.P. and J.N.M.G. performed the structural analysis of spike proteins. T.W. and D.F. performed and analyzed the SARS-CoV assays. J.Q.K. performed and analyzed IC 50 experiments. M.R. and J.W. provided SARS-CoV-2 replicon. I.M. provided the normal human bronchial epithelial cells. All authors approved the final version to be published.
DECLARATION OF INTERESTS The authors declare no competing interests.
STAR★METHODS Detailed methods are provided in the online version of this paper and include the following:
Viral molecular clones Viral clones were generated and produced as described in. 38 The mutant virus spike (SpikeΔ9) was generated by mutating the Fragment #8 using the Q5 Site-Directed Mutagenesis Kit (E0554) from New England Biolabs. Using the following primers: Forward Primer AGAGGTTTGATAACCCTG Reverse Primer TAGCATGGAACCAAGTAAC.
SARS-CoV-2 Replicon The CMV promoter driven SARS-CoV-2 delta variant subgenomic replicon was synthesized by Telesis Bio (formerly Codex DNA). Briefly, the replicon was designed by replacing Spike and envelope protein genes with cassettes for Zeocin resistance and nanoluciferase containing an IL-6 signal sequence for..
References
Abassi, Bertoglio, Ma� Cak � Safranko, Schirrmann, Greweling-Pils et al., Evaluation of the Neutralizing Antibody STE90-C11 against SARS-CoV-2 Delta Infection and Its Recognition of Other Variants of Concerns, Viruses
Abramson, Adler, Dunger, Evans, Green et al., Accurate structure prediction of biomolecular interactions with AlphaFold 3, Nature
Baggen, Persoons, Vanstreels, Jansen, Van Looveren et al., Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2, Nat. Genet
Barros De Lima, Nencioni, Thimoteo, Perea, Pinto et al., TMPRSS2 as a Key Player in Viral Pathogenesis: Influenza and Coronaviruses, Biomolecules
Biering, Sarnik, Wang, Zengel, Leist et al., Genome-wide bidirectional CRISPR screens identify mucins as host factors modulating SARS-CoV-2 infection, Nat. Genet
Bradley, Smith, Lee, Gupta, SARS-CoV-2 spike N-terminal domain modulates TMPRSS2-dependent viral entry and fusogenicity, Cell Rep
Buchrieser, Dufloo, Hubert, Monel, Planas et al., Syncytia formation by SARS-CoV-2-infected cells, EMBO J
Cao, Yisimayi, Bai, Huang, Li et al., Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines, Cell Res
Chan, Farias, Lee, Guvenc, Mero et al., Survival-based CRISPR genetic screens across a panel of permissive cell lines identify common and cell-specific SARS-CoV-2 host factors, Heliyon
Christian, Collier, Zu, Licursi, Hough et al., Activated Ras/MEK inhibits the antiviral response of alpha interferon by reducing STAT2 levels, J. Virol
Christian, Zu, Licursi, Komatsu, Pongnopparat et al., Suppression of IFN-induced transcription underlies IFN defects generated by activated Ras/MEK in human cancer cells, PLoS One
Deng, Atyeo, Yuan, Chicz, Tibbitts et al., Betaspike-containing boosters induce robust and functional antibody responses to SARS-CoV-2 in macaques primed with distinct vaccines, Cell Rep
Doench, Fusi, Sullender, Hegde, Vaimberg et al., Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9, Nat. Biotechnol
Flores-Martinez, Le-Trilling, Trilling, Nedd8-Activating Enzyme Is a Druggable Host Dependency Factor of Human and Mouse Cytomegalovirus, Viruses
Fu, Wang, Targeting NEDD8-activating enzyme for cancer therapy: developments, clinical trials, challenges and future research directions, J. Hematol. Oncol
Ghasemnejad-Berenji, Pashapour, SARS-CoV-2 and the Possible Role of Raf/MEK/ERK Pathway in Viral Survival: Is This a Potential Therapeutic Strategy for COVID-19?, Pharmacology
Grodzki, Bluhm, Schaefer, Tagmount, Russo et al., Genome-scale CRISPR screens identify host factors that promote human coronavirus infection, Genome Med
Gupta, Jayakumar, Saleh, Kannan, Halwani et al., SARS-CoV-2 infection-induced growth factors play differential roles in COVID-19 pathogenesis, Life Sci
Han, Zhang, Roles of neddylation against viral infections, Cell. Mol. Immunol
Harioudh, Perez, Chong, Nair, So et al., Oligoadenylate synthetase 1 displays dual antiviral mechanisms in driving translational shutdown and protecting interferon production, Immunity
Hornak, Abel, Okur, Strockbine, Roitberg et al., Comparison of multiple Amber force fields and development of improved protein backbone parameters, Proteins
Huang, Yang, Xu, Xu, Liu, Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19, Acta Pharmacol. Sin
Israeli, Finkel, Yahalom-Ronen, Paran, Chitlaru et al., Genomewide CRISPR screens identify GATA6 as a proviral host factor for SARS-CoV-2 via modulation of ACE2, Nat. Commun
Jackson, Farzan, Chen, Choe, Mechanisms of SARS-CoV-2 entry into cells, Nat. Rev. Mol. Cell Biol
Joung, Konermann, Gootenberg, Abudayyeh, Platt et al., Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening, Nat. Protoc
Jumper, Evans, Pritzel, Green, Figurnov et al., Highly accurate protein structure prediction with AlphaFold, Nature
Kakizaki, Hashimoto, Nagata, Yamamoto, Okura et al., The respective roles of TMPRSS2 and cathepsins for SARS-CoV-2 infection in human respiratory organoids, J. Virol
Kayesh, Kohara, Tsukiyama-Kohara, Effects of neddylation on viral infection: an overview, Arch. Virol
Kumar, Ishida, Strilets, Cole, Lopez-Orozco et al., SARS-CoV-2 Nonstructural Protein 1 Inhibits the Interferon Response by Causing Depletion of Key Host Signaling Factors, J. Virol
Le-Trilling, Megger, Katschinski, Landsberg, Ru ¨ Ckborn et al., Broad and potent antiviral activity of the NAE inhibitor MLN4924, Sci. Rep
Lee, Tebaldi, Pritchard, Nicola, Host Cell Neddylation Facilitates Alphaherpesvirus Entry in a Virus-Specific and Cell-Dependent Manner, Microbiol. Spectr
Leist, Dinnon, Scha ¨ Fer, Tse, Okuda et al., A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice, Cell
Li, Hilgenfeld, Whitley, De Clercq, Therapeutic strategies for COVID-19: progress and lessons learned, Nat. Rev. Drug Discov
Li, Xu, Xiao, Cong, Love et al., MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens, Genome Biol
Limburg, Harbig, Bestle, Stein, Moulton et al., TMPRSS2 Is the Major Activating Protease of Influenza A Virus in Primary Human Airway Cells and Influenza B Virus in Human Type II Pneumocytes, J. Virol
Limonta, Jovel, Kumar, Lu, Hou et al., Fibroblast Growth Factor 2 Enhances Zika Virus Infection in Human Fetal Brain, J. Infect. Dis
Liu, Zhang, Yang, Jiao, Zhao et al., HDM2 Promotes NEDDylation of Hepatitis B Virus HBx To Enhance Its Stability and Function, J. Virol
Lopez-Orozco, Fayad, Khan, Felix-Lopez, Elaish et al., The RNA Interference Effector Protein Argonaute 2 Functions as a Restriction Factor Against SARS-CoV-2, J. Mol. Biol
Ly, Ikegami, Rift Valley fever virus NSs protein functions and the similarity to other bunyavirus NSs proteins, Virol. J
Maddaluno, Urwyler, Rauschendorfer, Meyer, Stefanova et al., Antagonism of interferon signaling by fibroblast growth factors promotes viral replication, EMBO Mol. Med
Mandhana, Horvath, Sendai Virus Infection Induces Expression of Novel RNAs in Human Cells, Sci. Rep
Martinez-Gil, Goff, Garcı ´a-Sastre, Shaw, Palese, A Sendai virus-derived RNA agonist of RIG-I as a virus vaccine adjuvant, J. Virol
Meng, Datir, Choi, CITIID-NIHR Bioresource
Meng, Kemp, Papa, Datir, Ferreira et al., Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7, Cell Rep
Mirdita, Schu ¨ Tze, Moriwaki, Heo, Ovchinnikov et al., ColabFold: making protein folding accessible to all, Nat. Methods
Pavelin, Jin, Gratacap, Taggart, Hamilton et al., The nedd-8 activating enzyme gene underlies genetic resistance to infectious pancreatic necrosis virus in Atlantic salmon, Genomics
Pleschka, Wolff, Ehrhardt, Hobom, Planz et al., Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade, Nat. Cell Biol
Pormohammad, Zarei, Ghorbani, Mohammadi, Sheikh Neshin et al., Effectiveness of COVID-19 Vaccines against Delta (B.1.617.2) Variant: A Systematic Review and Meta-Analysis of Clinical Studies, Vaccines
Qing, Kicmal, Kumar, Hawkins, Timm et al., Dynamics of SARS-CoV-2 Spike Proteins in Cell Entry: Control Elements in the Amino-Terminal Domains, mBio
Qing, Li, Cooper, Schulz, Ja ¨ Ck et al., Inter-domain communication in SARS-CoV-2 spike proteins controls protease-triggered cell entry, Cell Rep
Qu, Nebioglu, Leuthold, Ni, Mutz et al., Dual role of neddylation in transcription of hepatitis B virus RNAs from cccDNA and production of viral surface antigen, JHEP Rep
Rebendenne, Roy, Bonaventure, Chaves Valada ˜ O, Desmarets et al., Bidirectional genome-wide CRISPR screens reveal host factors regulating SARS-CoV-2, MERS-CoV and seasonal HCoVs, Nat. Genet
Ren, Fu, Zhang, Ju, Jiang et al., Zika virus NS5 protein inhibits type I interferon signaling via CRL3 E3 ubiquitin ligase-mediated degradation of STAT2, Proc. Natl. Acad. Sci
Richard, Michael, Alexander, Natasha, Andrew et al., Protein complex prediction with AlphaFold-Multimer, doi:10.1101/2021.10.04.463034
Schneider, Luna, Hoffmann, Sa ´ Nchez-Rivera, Leal et al., Genome-Scale Identification of SARS-CoV-2 and Pan-coronavirus Host Factor Networks, Cell
Schrodinger, The PyMOL Molecular Graphics System
Shapira, Monreal, Dion, Buchholz, Imbiakha et al., A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic, Nature
Shen, Mao, Wu, Tanaka, Zhang, TMPRSS2: A potential target for treatment of influenza virus and coronavirus infections, Biochimie
Siwak, Leblanc, Scott, Kim, Pellizzari-Delano et al., Cellular sialoglycans are differentially required for endosomal and cell-surface entry of SARS-CoV-2 in lung cell lines, PLoS Pathog
Song, Huai, Yu, Wang, Zhao et al., MLN4924, a First-in-Class NEDD8-Activating Enzyme Inhibitor, Attenuates IFN-beta Production, J. Immunol
Subbiah, Verstovsek, Clinical development and management of adverse events associated with FGFR inhibitors, Cell Rep. Med
Sun, Yao, Wang, Qian, Chen et al., Inhibition of neddylation pathway represses influenza virus replication and pro-inflammatory responses, Virology
Taha, Irene, Jennifer, Takako, Keith et al., Rapid assembly of SARS-CoV-2 genomes reveals attenuation of the Omicron BA.1 variant through NSP6, doi:10.1101/2023.01.31.525914
Van Cleemput, Van Snippenberg, Lambrechts, Dendooven, D'onofrio et al., Organ-specific genome diversity of replicationcompetent SARS-CoV-2, Nat. Commun
Verhelst, Parthoens, Schepens, Fiers, Saelens, Interferon-inducible protein Mx1 inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly, J. Virol
Villenave, Touzelet, Thavagnanam, Sarlang, Parker et al., Cytopathogenesis of Sendai virus in well-differentiated primary pediatric bronchial epithelial cells, J. Virol
Xie, Su, Yang, Tan, Huang et al., FGF/FGFR signaling in health and disease, Signal Transduct. Target. Ther
Xu, Elaish, Wong, Hassan, Lopez-Orozco et al., The Wnt/β-catenin pathway is important for replication of SARS-CoV-2 and other pathogenic RNA viruses, npj Viruses
Xu, Shu, Kang, Wu, Zhou et al., Temporal profiling of plasma cytokines, chemokines and growth factors from mild, severe and fatal COVID-19 patients, Signal Transduct. Target. Ther
Zhang, Ye, Pei, Qiu, Wang et al., Neddylation is required for herpes simplex virus type I (HSV-1)induced early phase interferon-beta production, Cell. Mol. Immunol
Zhang, Yu, Li, Zhao, Sun, Protein neddylation and its role in health and diseases, Signal Transduct. Target. Ther
Zhu, Feng, Hu, Wang, Yu et al., A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry, Nat. Commun
DOI record:
{
"DOI": "10.1016/j.isci.2025.114566",
"ISSN": [
"2589-0042"
],
"URL": "http://dx.doi.org/10.1016/j.isci.2025.114566",
"alternative-id": [
"S2589004225028275"
],
"article-number": "114566",
"assertion": [
{
"label": "This article is maintained by",
"name": "publisher",
"value": "Elsevier"
},
{
"label": "Article Title",
"name": "articletitle",
"value": "FGFR signaling and neddylation facilitate SARS-CoV-2 infection by modulating interferon induction and viral entry, respectively"
},
{
"label": "Journal Title",
"name": "journaltitle",
"value": "iScience"
},
{
"label": "CrossRef DOI link to publisher maintained version",
"name": "articlelink",
"value": "https://doi.org/10.1016/j.isci.2025.114566"
},
{
"label": "Content Type",
"name": "content_type",
"value": "article"
},
{
"label": "Copyright",
"name": "copyright",
"value": "© 2025 The Author(s). Published by Elsevier Inc."
}
],
"author": [
{
"affiliation": [],
"family": "Felix-Lopez",
"given": "Alberto",
"sequence": "first"
},
{
"affiliation": [],
"family": "Lopez-Orozco",
"given": "Joaquin",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Elaish",
"given": "Mohamed",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Fayad",
"given": "Nawell",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Xu",
"given": "Zaikun",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Woldemariam",
"given": "Tekeleselassie",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hassan",
"given": "Bardes B.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Panigrahi",
"given": "Rashmi",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Khan",
"given": "Juveriya Qamar",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Rohamare",
"given": "Megha",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Mayers",
"given": "Irv",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Glover",
"given": "J.N. Mark",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wilson",
"given": "Joyce A.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Falzarano",
"given": "Darryl",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Kumar",
"given": "Anil",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-5744-3545",
"affiliation": [],
"authenticated-orcid": false,
"family": "Hobman",
"given": "Tom C.",
"sequence": "additional"
}
],
"container-title": "iScience",
"container-title-short": "iScience",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"cell.com",
"elsevier.com",
"sciencedirect.com"
]
},
"created": {
"date-parts": [
[
2025,
12,
29
]
],
"date-time": "2025-12-29T07:24:52Z",
"timestamp": 1766993092000
},
"deposited": {
"date-parts": [
[
2026,
1,
21
]
],
"date-time": "2026-01-21T16:10:45Z",
"timestamp": 1769011845000
},
"funder": [
{
"DOI": "10.13039/100013200",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100013200",
"id-type": "DOI"
}
],
"name": "Government of Alberta"
},
{
"DOI": "10.13039/501100017001",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100017001",
"id-type": "DOI"
}
],
"name": "Li Ka Shing Institute of Virology, University of Alberta"
},
{
"DOI": "10.13039/501100000024",
"award": [
"GA1-177707"
],
"award-info": [
{
"award-number": [
"GA1-177707"
]
}
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100000024",
"id-type": "DOI"
}
],
"name": "Canadian Institutes of Health Research"
},
{
"DOI": "10.13039/501100000024",
"award": [
"VR3-172626"
],
"award-info": [
{
"award-number": [
"VR3-172626"
]
}
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100000024",
"id-type": "DOI"
}
],
"name": "Canadian Institutes of Health Research"
},
{
"DOI": "10.13039/501100000024",
"award": [
"PUU-177963"
],
"award-info": [
{
"award-number": [
"PUU-177963"
]
}
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100000024",
"id-type": "DOI"
}
],
"name": "Canadian Institutes of Health Research"
},
{
"DOI": "10.13039/501100000024",
"award": [
"MM1-181121"
],
"award-info": [
{
"award-number": [
"MM1-181121"
]
}
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100000024",
"id-type": "DOI"
}
],
"name": "Canadian Institutes of Health Research"
},
{
"DOI": "10.13039/501100000024",
"award": [
"PTT-179810"
],
"award-info": [
{
"award-number": [
"PTT-179810"
]
}
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100000024",
"id-type": "DOI"
}
],
"name": "Canadian Institutes of Health Research"
},
{
"DOI": "10.13039/501100000190",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100000190",
"id-type": "DOI"
}
],
"name": "University of Alberta"
},
{
"DOI": "10.13039/100021824",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100021824",
"id-type": "DOI"
}
],
"name": "Government of Saskatchewan"
},
{
"DOI": "10.13039/100008920",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100008920",
"id-type": "DOI"
}
],
"name": "University of Saskatchewan"
},
{
"DOI": "10.13039/501100000196",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100000196",
"id-type": "DOI"
}
],
"name": "Canada Foundation for Innovation"
}
],
"indexed": {
"date-parts": [
[
2026,
1,
22
]
],
"date-time": "2026-01-22T07:28:34Z",
"timestamp": 1769066914816,
"version": "3.49.0"
},
"is-referenced-by-count": 0,
"issue": "2",
"issued": {
"date-parts": [
[
2026,
2
]
]
},
"journal-issue": {
"issue": "2",
"published-print": {
"date-parts": [
[
2026,
2
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://www.elsevier.com/tdm/userlicense/1.0/",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2026,
2,
1
]
],
"date-time": "2026-02-01T00:00:00Z",
"timestamp": 1769904000000
}
},
{
"URL": "https://www.elsevier.com/legal/tdmrep-license",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2026,
2,
1
]
],
"date-time": "2026-02-01T00:00:00Z",
"timestamp": 1769904000000
}
},
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
12,
23
]
],
"date-time": "2025-12-23T00:00:00Z",
"timestamp": 1766448000000
}
}
],
"link": [
{
"URL": "https://api.elsevier.com/content/article/PII:S2589004225028275?httpAccept=text/xml",
"content-type": "text/xml",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://api.elsevier.com/content/article/PII:S2589004225028275?httpAccept=text/plain",
"content-type": "text/plain",
"content-version": "vor",
"intended-application": "text-mining"
}
],
"member": "78",
"original-title": [],
"page": "114566",
"prefix": "10.1016",
"published": {
"date-parts": [
[
2026,
2
]
]
},
"published-print": {
"date-parts": [
[
2026,
2
]
]
},
"publisher": "Elsevier BV",
"reference": [
{
"DOI": "10.1038/s41467-021-26884-7",
"article-title": "Organ-specific genome diversity of replication-competent SARS-CoV-2",
"author": "Van Cleemput",
"doi-asserted-by": "crossref",
"first-page": "6612",
"journal-title": "Nat. Commun.",
"key": "10.1016/j.isci.2025.114566_bib1",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1038/s41564-020-0695-z",
"article-title": "The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2",
"doi-asserted-by": "crossref",
"first-page": "536",
"journal-title": "Nat. Microbiol.",
"key": "10.1016/j.isci.2025.114566_bib2",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.1016/j.celrep.2023.113292",
"article-title": "Beta-spike-containing boosters induce robust and functional antibody responses to SARS-CoV-2 in macaques primed with distinct vaccines",
"author": "Deng",
"doi-asserted-by": "crossref",
"journal-title": "Cell Rep.",
"key": "10.1016/j.isci.2025.114566_bib3",
"volume": "42",
"year": "2023"
},
{
"DOI": "10.3390/v15112153",
"article-title": "Evaluation of the Neutralizing Antibody STE90-C11 against SARS-CoV-2 Delta Infection and Its Recognition of Other Variants of Concerns",
"author": "Abassi",
"doi-asserted-by": "crossref",
"journal-title": "Viruses",
"key": "10.1016/j.isci.2025.114566_bib4",
"volume": "15",
"year": "2023"
},
{
"article-title": "Effectiveness of COVID-19 Vaccines against Delta (B.1.617.2) Variant: A Systematic Review and Meta-Analysis of Clinical Studies",
"author": "Pormohammad",
"journal-title": "Vaccines (Basel)",
"key": "10.1016/j.isci.2025.114566_bib5",
"volume": "10",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.12.006",
"article-title": "Genome-Scale Identification of SARS-CoV-2 and Pan-coronavirus Host Factor Networks",
"author": "Schneider",
"doi-asserted-by": "crossref",
"first-page": "120",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.114566_bib6",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1038/s41588-022-01110-2",
"article-title": "Bidirectional genome-wide CRISPR screens reveal host factors regulating SARS-CoV-2, MERS-CoV and seasonal HCoVs",
"author": "Rebendenne",
"doi-asserted-by": "crossref",
"first-page": "1090",
"journal-title": "Nat. Genet.",
"key": "10.1016/j.isci.2025.114566_bib7",
"volume": "54",
"year": "2022"
},
{
"DOI": "10.1038/s41588-021-00805-2",
"article-title": "Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2",
"author": "Baggen",
"doi-asserted-by": "crossref",
"first-page": "435",
"journal-title": "Nat. Genet.",
"key": "10.1016/j.isci.2025.114566_bib8",
"volume": "53",
"year": "2021"
},
{
"DOI": "10.1186/s13073-022-01013-1",
"article-title": "Genome-scale CRISPR screens identify host factors that promote human coronavirus infection",
"author": "Grodzki",
"doi-asserted-by": "crossref",
"first-page": "10",
"journal-title": "Genome Med.",
"key": "10.1016/j.isci.2025.114566_bib9",
"volume": "14",
"year": "2022"
},
{
"DOI": "10.1038/s44298-024-00018-4",
"article-title": "The Wnt/β-catenin pathway is important for replication of SARS-CoV-2 and other pathogenic RNA viruses",
"author": "Xu",
"doi-asserted-by": "crossref",
"first-page": "6",
"journal-title": "npj Viruses",
"key": "10.1016/j.isci.2025.114566_bib10",
"volume": "2",
"year": "2024"
},
{
"DOI": "10.1016/j.cell.2020.09.050",
"article-title": "A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice",
"author": "Leist",
"doi-asserted-by": "crossref",
"first-page": "1070",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.114566_bib11",
"volume": "183",
"year": "2020"
},
{
"DOI": "10.1128/jvi.01853-24",
"article-title": "The respective roles of TMPRSS2 and cathepsins for SARS-CoV-2 infection in human respiratory organoids",
"author": "Kakizaki",
"doi-asserted-by": "crossref",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.114566_bib12",
"volume": "99",
"year": "2025"
},
{
"DOI": "10.3390/biom15010075",
"article-title": "TMPRSS2 as a Key Player in Viral Pathogenesis: Influenza and Coronaviruses",
"author": "Barros de Lima",
"doi-asserted-by": "crossref",
"journal-title": "Biomolecules",
"key": "10.1016/j.isci.2025.114566_bib13",
"volume": "15",
"year": "2025"
},
{
"DOI": "10.1038/nbt.3437",
"article-title": "Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9",
"author": "Doench",
"doi-asserted-by": "crossref",
"first-page": "184",
"journal-title": "Nat. Biotechnol.",
"key": "10.1016/j.isci.2025.114566_bib14",
"volume": "34",
"year": "2016"
},
{
"DOI": "10.1128/JVI.00266-21",
"article-title": "SARS-CoV-2 Nonstructural Protein 1 Inhibits the Interferon Response by Causing Depletion of Key Host Signaling Factors",
"author": "Kumar",
"doi-asserted-by": "crossref",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.114566_bib15",
"volume": "95",
"year": "2021"
},
{
"DOI": "10.1038/nprot.2017.016",
"article-title": "Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening",
"author": "Joung",
"doi-asserted-by": "crossref",
"first-page": "828",
"journal-title": "Nat. Protoc.",
"key": "10.1016/j.isci.2025.114566_bib16",
"volume": "12",
"year": "2017"
},
{
"DOI": "10.1186/s13059-014-0554-4",
"article-title": "MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "554",
"journal-title": "Genome Biol.",
"key": "10.1016/j.isci.2025.114566_bib17",
"volume": "15",
"year": "2014"
},
{
"DOI": "10.1038/s41467-021-21213-4",
"article-title": "A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry",
"author": "Zhu",
"doi-asserted-by": "crossref",
"first-page": "961",
"journal-title": "Nat. Commun.",
"key": "10.1016/j.isci.2025.114566_bib18",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1016/j.heliyon.2022.e12744",
"article-title": "Survival-based CRISPR genetic screens across a panel of permissive cell lines identify common and cell-specific SARS-CoV-2 host factors",
"author": "Chan",
"doi-asserted-by": "crossref",
"journal-title": "Heliyon",
"key": "10.1016/j.isci.2025.114566_bib19",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.1038/s41392-020-00222-7",
"article-title": "FGF/FGFR signaling in health and disease",
"author": "Xie",
"doi-asserted-by": "crossref",
"first-page": "181",
"journal-title": "Signal Transduct. Target. Ther.",
"key": "10.1016/j.isci.2025.114566_bib20",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.1159/000511280",
"article-title": "SARS-CoV-2 and the Possible Role of Raf/MEK/ERK Pathway in Viral Survival: Is This a Potential Therapeutic Strategy for COVID-19?",
"author": "Ghasemnejad-Berenji",
"doi-asserted-by": "crossref",
"first-page": "119",
"journal-title": "Pharmacology",
"key": "10.1016/j.isci.2025.114566_bib21",
"volume": "106",
"year": "2021"
},
{
"DOI": "10.1093/infdis/jiz073",
"article-title": "Fibroblast Growth Factor 2 Enhances Zika Virus Infection in Human Fetal Brain",
"author": "Limonta",
"doi-asserted-by": "crossref",
"first-page": "1377",
"journal-title": "J. Infect. Dis.",
"key": "10.1016/j.isci.2025.114566_bib22",
"volume": "220",
"year": "2019"
},
{
"DOI": "10.1038/35060098",
"article-title": "Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade",
"author": "Pleschka",
"doi-asserted-by": "crossref",
"first-page": "301",
"journal-title": "Nat. Cell Biol.",
"key": "10.1016/j.isci.2025.114566_bib23",
"volume": "3",
"year": "2001"
},
{
"DOI": "10.1128/JVI.02213-08",
"article-title": "Activated Ras/MEK inhibits the antiviral response of alpha interferon by reducing STAT2 levels",
"author": "Christian",
"doi-asserted-by": "crossref",
"first-page": "6717",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.114566_bib24",
"volume": "83",
"year": "2009"
},
{
"DOI": "10.1128/JVI.02338-12",
"article-title": "A Sendai virus-derived RNA agonist of RIG-I as a virus vaccine adjuvant",
"author": "Martinez-Gil",
"doi-asserted-by": "crossref",
"first-page": "1290",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.114566_bib25",
"volume": "87",
"year": "2013"
},
{
"DOI": "10.1128/JVI.00798-10",
"article-title": "Cytopathogenesis of Sendai virus in well-differentiated primary pediatric bronchial epithelial cells",
"author": "Villenave",
"doi-asserted-by": "crossref",
"first-page": "11718",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.114566_bib26",
"volume": "84",
"year": "2010"
},
{
"DOI": "10.1038/s41598-018-35231-8",
"article-title": "Sendai Virus Infection Induces Expression of Novel RNAs in Human Cells",
"author": "Mandhana",
"doi-asserted-by": "crossref",
"journal-title": "Sci. Rep.",
"key": "10.1016/j.isci.2025.114566_bib27",
"volume": "8",
"year": "2018"
},
{
"DOI": "10.1371/journal.pone.0044267",
"article-title": "Suppression of IFN-induced transcription underlies IFN defects generated by activated Ras/MEK in human cancer cells",
"author": "Christian",
"doi-asserted-by": "crossref",
"journal-title": "PLoS One",
"key": "10.1016/j.isci.2025.114566_bib28",
"volume": "7",
"year": "2012"
},
{
"DOI": "10.1038/cmi.2017.100",
"article-title": "Roles of neddylation against viral infections",
"author": "Han",
"doi-asserted-by": "crossref",
"first-page": "292",
"journal-title": "Cell. Mol. Immunol.",
"key": "10.1016/j.isci.2025.114566_bib29",
"volume": "15",
"year": "2018"
},
{
"DOI": "10.4049/jimmunol.1501752",
"article-title": "MLN4924, a First-in-Class NEDD8-Activating Enzyme Inhibitor, Attenuates IFN-beta Production",
"author": "Song",
"doi-asserted-by": "crossref",
"first-page": "3117",
"journal-title": "J. Immunol.",
"key": "10.1016/j.isci.2025.114566_bib30",
"volume": "196",
"year": "2016"
},
{
"DOI": "10.1038/s41392-024-01800-9",
"article-title": "Protein neddylation and its role in health and diseases",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "85",
"journal-title": "Signal Transduct. Target. Ther.",
"key": "10.1016/j.isci.2025.114566_bib31",
"volume": "9",
"year": "2024"
},
{
"DOI": "10.1007/s00705-023-05930-3",
"article-title": "Effects of neddylation on viral infection: an overview",
"author": "Kayesh",
"doi-asserted-by": "crossref",
"first-page": "6",
"journal-title": "Arch. Virol.",
"key": "10.1016/j.isci.2025.114566_bib32",
"volume": "169",
"year": "2023"
},
{
"DOI": "10.1038/srep19977",
"article-title": "Broad and potent antiviral activity of the NAE inhibitor MLN4924",
"author": "Le-Trilling",
"doi-asserted-by": "crossref",
"journal-title": "Sci. Rep.",
"key": "10.1016/j.isci.2025.114566_bib33",
"volume": "6",
"year": "2016"
},
{
"DOI": "10.1016/j.virol.2017.11.004",
"article-title": "Inhibition of neddylation pathway represses influenza virus replication and pro-inflammatory responses",
"author": "Sun",
"doi-asserted-by": "crossref",
"first-page": "230",
"journal-title": "Virology",
"key": "10.1016/j.isci.2025.114566_bib34",
"volume": "514",
"year": "2018"
},
{
"DOI": "10.1073/pnas.2403235121",
"article-title": "Zika virus NS5 protein inhibits type I interferon signaling via CRL3 E3 ubiquitin ligase-mediated degradation of STAT2",
"author": "Ren",
"doi-asserted-by": "crossref",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "10.1016/j.isci.2025.114566_bib35",
"volume": "121",
"year": "2024"
},
{
"DOI": "10.1128/JVI.00340-17",
"article-title": "HDM2 Promotes NEDDylation of Hepatitis B Virus HBx To Enhance Its Stability and Function",
"author": "Liu",
"doi-asserted-by": "crossref",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.114566_bib36",
"volume": "91",
"year": "2017"
},
{
"DOI": "10.1038/s41401-020-0485-4",
"article-title": "Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19",
"author": "Huang",
"doi-asserted-by": "crossref",
"first-page": "1141",
"journal-title": "Acta Pharmacol. Sin.",
"key": "10.1016/j.isci.2025.114566_bib37",
"volume": "41",
"year": "2020"
},
{
"article-title": "Rapid assembly of SARS-CoV-2 genomes reveals attenuation of the Omicron BA.1 variant through NSP6",
"author": "Taha",
"journal-title": "bioRxiv",
"key": "10.1016/j.isci.2025.114566_bib38",
"year": "2023"
},
{
"DOI": "10.1038/s41422-021-00514-9",
"article-title": "Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines",
"author": "Cao",
"doi-asserted-by": "crossref",
"first-page": "732",
"journal-title": "Cell Res.",
"key": "10.1016/j.isci.2025.114566_bib39",
"volume": "31",
"year": "2021"
},
{
"DOI": "10.1128/spectrum.03114-22",
"article-title": "Host Cell Neddylation Facilitates Alphaherpesvirus Entry in a Virus-Specific and Cell-Dependent Manner",
"author": "Lee",
"doi-asserted-by": "crossref",
"journal-title": "Microbiol. Spectr.",
"key": "10.1016/j.isci.2025.114566_bib40",
"volume": "10",
"year": "2022"
},
{
"DOI": "10.1038/s41580-021-00418-x",
"article-title": "Mechanisms of SARS-CoV-2 entry into cells",
"author": "Jackson",
"doi-asserted-by": "crossref",
"first-page": "3",
"journal-title": "Nat. Rev. Mol. Cell Biol.",
"key": "10.1016/j.isci.2025.114566_bib41",
"volume": "23",
"year": "2022"
},
{
"DOI": "10.1371/journal.ppat.1012365",
"article-title": "Cellular sialoglycans are differentially required for endosomal and cell-surface entry of SARS-CoV-2 in lung cell lines",
"author": "Siwak",
"doi-asserted-by": "crossref",
"journal-title": "PLoS Pathog.",
"key": "10.1016/j.isci.2025.114566_bib42",
"volume": "20",
"year": "2024"
},
{
"DOI": "10.1016/j.celrep.2022.111220",
"article-title": "SARS-CoV-2 spike N-terminal domain modulates TMPRSS2-dependent viral entry and fusogenicity",
"author": "Meng",
"doi-asserted-by": "crossref",
"journal-title": "Cell Rep.",
"key": "10.1016/j.isci.2025.114566_bib43",
"volume": "40",
"year": "2022"
},
{
"DOI": "10.1016/j.celrep.2021.109292",
"article-title": "Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7",
"author": "Meng",
"doi-asserted-by": "crossref",
"journal-title": "Cell Rep.",
"key": "10.1016/j.isci.2025.114566_bib44",
"volume": "35",
"year": "2021"
},
{
"DOI": "10.15252/embj.2020106267",
"article-title": "Syncytia formation by SARS-CoV-2-infected cells",
"author": "Buchrieser",
"doi-asserted-by": "crossref",
"journal-title": "EMBO J.",
"key": "10.1016/j.isci.2025.114566_bib45",
"volume": "39",
"year": "2020"
},
{
"DOI": "10.1128/mBio.01590-21",
"article-title": "Dynamics of SARS-CoV-2 Spike Proteins in Cell Entry: Control Elements in the Amino-Terminal Domains",
"author": "Qing",
"doi-asserted-by": "crossref",
"journal-title": "mBio",
"key": "10.1016/j.isci.2025.114566_bib46",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1016/j.celrep.2022.110786",
"article-title": "Inter-domain communication in SARS-CoV-2 spike proteins controls protease-triggered cell entry",
"author": "Qing",
"doi-asserted-by": "crossref",
"journal-title": "Cell Rep.",
"key": "10.1016/j.isci.2025.114566_bib47",
"volume": "39",
"year": "2022"
},
{
"DOI": "10.1128/JVI.00649-19",
"article-title": "TMPRSS2 Is the Major Activating Protease of Influenza A Virus in Primary Human Airway Cells and Influenza B Virus in Human Type II Pneumocytes",
"author": "Limburg",
"doi-asserted-by": "crossref",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.114566_bib48",
"volume": "93",
"year": "2019"
},
{
"DOI": "10.1038/s41586-022-04661-w",
"article-title": "A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic",
"author": "Shapira",
"doi-asserted-by": "crossref",
"first-page": "340",
"journal-title": "Nature",
"key": "10.1016/j.isci.2025.114566_bib49",
"volume": "605",
"year": "2022"
},
{
"DOI": "10.1016/j.biochi.2017.07.016",
"article-title": "TMPRSS2: A potential target for treatment of influenza virus and coronavirus infections",
"author": "Shen",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "Biochimie",
"key": "10.1016/j.isci.2025.114566_bib50",
"volume": "142",
"year": "2017"
},
{
"DOI": "10.1038/cmi.2015.35",
"article-title": "Neddylation is required for herpes simplex virus type I (HSV-1)-induced early phase interferon-beta production",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "578",
"journal-title": "Cell. Mol. Immunol.",
"key": "10.1016/j.isci.2025.114566_bib51",
"volume": "13",
"year": "2016"
},
{
"DOI": "10.1038/s41588-022-01131-x",
"article-title": "Genome-wide bidirectional CRISPR screens identify mucins as host factors modulating SARS-CoV-2 infection",
"author": "Biering",
"doi-asserted-by": "crossref",
"first-page": "1078",
"journal-title": "Nat. Genet.",
"key": "10.1016/j.isci.2025.114566_bib52",
"volume": "54",
"year": "2022"
},
{
"DOI": "10.1038/s41467-022-29896-z",
"article-title": "Genome-wide CRISPR screens identify GATA6 as a proviral host factor for SARS-CoV-2 via modulation of ACE2",
"author": "Israeli",
"doi-asserted-by": "crossref",
"first-page": "2237",
"journal-title": "Nat. Commun.",
"key": "10.1016/j.isci.2025.114566_bib53",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1016/j.immuni.2024.02.002",
"article-title": "Oligoadenylate synthetase 1 displays dual antiviral mechanisms in driving translational shutdown and protecting interferon production",
"author": "Harioudh",
"doi-asserted-by": "crossref",
"first-page": "446",
"journal-title": "Immunity",
"key": "10.1016/j.isci.2025.114566_bib54",
"volume": "57",
"year": "2024"
},
{
"DOI": "10.1128/JVI.01682-12",
"article-title": "Interferon-inducible protein Mx1 inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly",
"author": "Verhelst",
"doi-asserted-by": "crossref",
"first-page": "13445",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.114566_bib55",
"volume": "86",
"year": "2012"
},
{
"DOI": "10.15252/emmm.201911793",
"article-title": "Antagonism of interferon signaling by fibroblast growth factors promotes viral replication",
"author": "Maddaluno",
"doi-asserted-by": "crossref",
"journal-title": "EMBO Mol. Med.",
"key": "10.1016/j.isci.2025.114566_bib56",
"volume": "12",
"year": "2020"
},
{
"DOI": "10.1038/s41392-020-0211-1",
"article-title": "Temporal profiling of plasma cytokines, chemokines and growth factors from mild, severe and fatal COVID-19 patients",
"author": "Xu",
"doi-asserted-by": "crossref",
"first-page": "100",
"journal-title": "Signal Transduct. Target. Ther.",
"key": "10.1016/j.isci.2025.114566_bib57",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.1016/j.lfs.2022.120703",
"article-title": "SARS-CoV-2 infection- induced growth factors play differential roles in COVID-19 pathogenesis",
"author": "Gupta",
"doi-asserted-by": "crossref",
"journal-title": "Life Sci.",
"key": "10.1016/j.isci.2025.114566_bib58",
"volume": "304",
"year": "2022"
},
{
"DOI": "10.3390/v13081610",
"article-title": "Nedd8-Activating Enzyme Is a Druggable Host Dependency Factor of Human and Mouse Cytomegalovirus",
"author": "Flores-Martinez",
"doi-asserted-by": "crossref",
"journal-title": "Viruses",
"key": "10.1016/j.isci.2025.114566_bib59",
"volume": "13",
"year": "2021"
},
{
"article-title": "Dual role of neddylation in transcription of hepatitis B virus RNAs from cccDNA and production of viral surface antigen",
"author": "Qu",
"journal-title": "JHEP Rep.",
"key": "10.1016/j.isci.2025.114566_bib60",
"volume": "4",
"year": "2022"
},
{
"DOI": "10.1186/s12985-016-0573-8",
"article-title": "Rift Valley fever virus NSs protein functions and the similarity to other bunyavirus NSs proteins",
"author": "Ly",
"doi-asserted-by": "crossref",
"first-page": "118",
"journal-title": "Virol. J.",
"key": "10.1016/j.isci.2025.114566_bib61",
"volume": "13",
"year": "2016"
},
{
"DOI": "10.1016/j.ygeno.2021.09.012",
"article-title": "The nedd-8 activating enzyme gene underlies genetic resistance to infectious pancreatic necrosis virus in Atlantic salmon",
"author": "Pavelin",
"doi-asserted-by": "crossref",
"first-page": "3842",
"journal-title": "Genomics",
"key": "10.1016/j.isci.2025.114566_bib62",
"volume": "113",
"year": "2021"
},
{
"article-title": "Clinical development and management of adverse events associated with FGFR inhibitors",
"author": "Subbiah",
"journal-title": "Cell Rep. Med.",
"key": "10.1016/j.isci.2025.114566_bib63",
"volume": "4",
"year": "2023"
},
{
"DOI": "10.1186/s13045-023-01485-7",
"article-title": "Targeting NEDD8-activating enzyme for cancer therapy: developments, clinical trials, challenges and future research directions",
"author": "Fu",
"doi-asserted-by": "crossref",
"first-page": "87",
"journal-title": "J. Hematol. Oncol.",
"key": "10.1016/j.isci.2025.114566_bib64",
"volume": "16",
"year": "2023"
},
{
"DOI": "10.1038/s41573-023-00672-y",
"article-title": "Therapeutic strategies for COVID-19: progress and lessons learned",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "449",
"journal-title": "Nat. Rev. Drug Discov.",
"key": "10.1016/j.isci.2025.114566_bib65",
"volume": "22",
"year": "2023"
},
{
"DOI": "10.1016/j.jmb.2023.168170",
"article-title": "The RNA Interference Effector Protein Argonaute 2 Functions as a Restriction Factor Against SARS-CoV-2",
"author": "Lopez-Orozco",
"doi-asserted-by": "crossref",
"journal-title": "J. Mol. Biol.",
"key": "10.1016/j.isci.2025.114566_bib66",
"volume": "435",
"year": "2023"
},
{
"DOI": "10.1038/s41592-022-01488-1",
"article-title": "ColabFold: making protein folding accessible to all",
"author": "Mirdita",
"doi-asserted-by": "crossref",
"first-page": "679",
"journal-title": "Nat. Methods",
"key": "10.1016/j.isci.2025.114566_bib67",
"volume": "19",
"year": "2022"
},
{
"DOI": "10.1038/s41586-021-03819-2",
"article-title": "Highly accurate protein structure prediction with AlphaFold",
"author": "Jumper",
"doi-asserted-by": "crossref",
"first-page": "583",
"journal-title": "Nature",
"key": "10.1016/j.isci.2025.114566_bib68",
"volume": "596",
"year": "2021"
},
{
"article-title": "Protein complex prediction with AlphaFold-Multimer",
"author": "Richard",
"journal-title": "bioRxiv",
"key": "10.1016/j.isci.2025.114566_bib69",
"year": "2022"
},
{
"DOI": "10.1002/prot.21123",
"article-title": "Comparison of multiple Amber force fields and development of improved protein backbone parameters",
"author": "Hornak",
"doi-asserted-by": "crossref",
"first-page": "712",
"journal-title": "Proteins",
"key": "10.1016/j.isci.2025.114566_bib70",
"volume": "65",
"year": "2006"
},
{
"author": "Schrodinger",
"key": "10.1016/j.isci.2025.114566_bib71"
},
{
"DOI": "10.1038/s41586-024-07487-w",
"article-title": "Accurate structure prediction of biomolecular interactions with AlphaFold 3",
"author": "Abramson",
"doi-asserted-by": "crossref",
"first-page": "493",
"journal-title": "Nature",
"key": "10.1016/j.isci.2025.114566_bib72",
"volume": "630",
"year": "2024"
}
],
"reference-count": 72,
"references-count": 72,
"relation": {},
"resource": {
"primary": {
"URL": "https://linkinghub.elsevier.com/retrieve/pii/S2589004225028275"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "FGFR signaling and neddylation facilitate SARS-CoV-2 infection by modulating interferon induction and viral entry, respectively",
"type": "journal-article",
"update-policy": "https://doi.org/10.1016/elsevier_cm_policy",
"volume": "29"
}