Intrahost evolution leading to distinct lineages in the upper and lower respiratory tracts during SARS-CoV-2 prolonged infection
et al., Virus Evolution, doi:10.1093/ve/veae073, Aug 2024
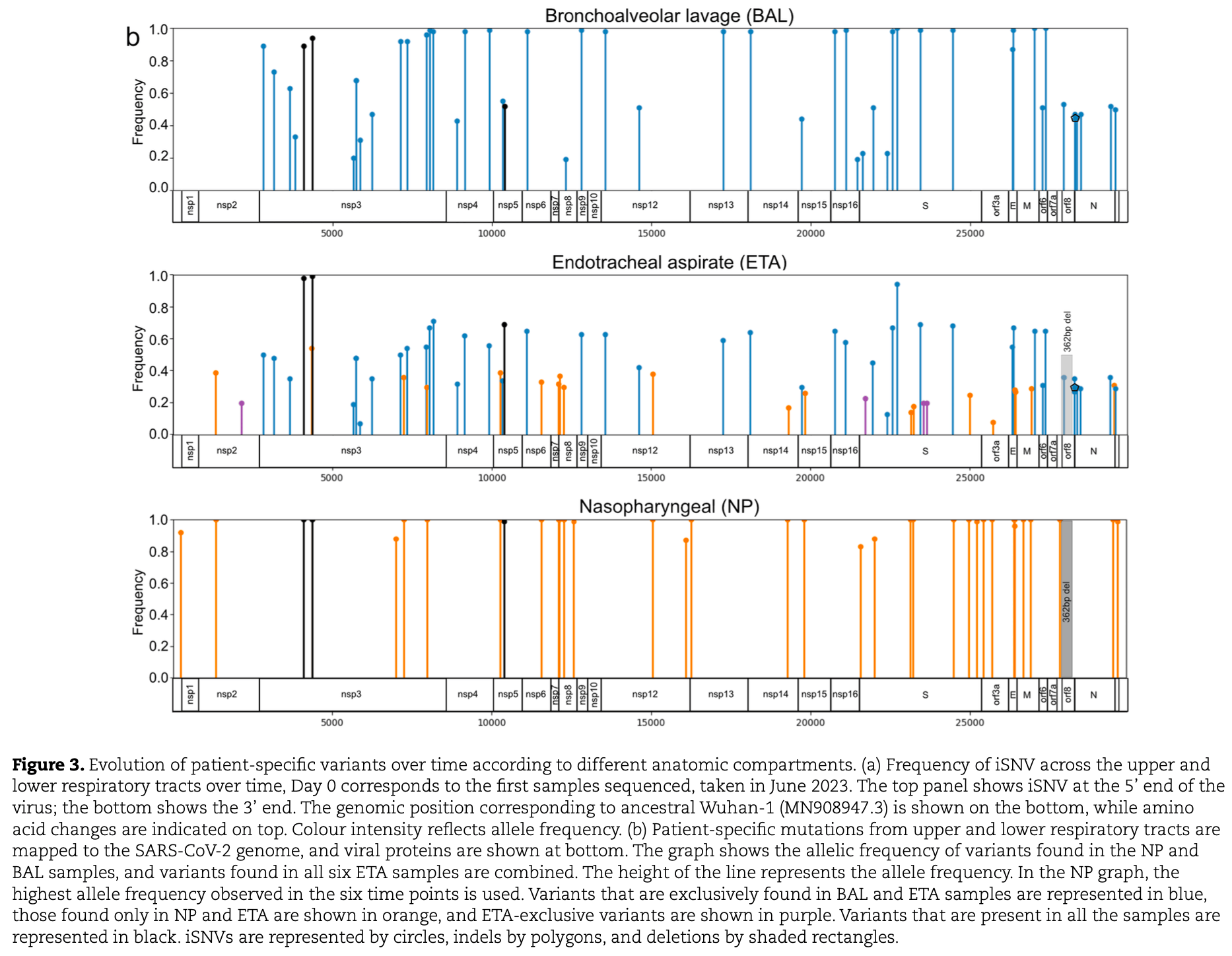
Genomic sequencing study of persistent SARS-CoV-2 infection in an immunocompromised individual showing genetically distinct viral lineages in the upper and lower respiratory tracts persisting for over 1 year. Sequencing revealed 87 intrahost single-nucleotide variants, 2 indels, and a 362-bp deletion, with nasopharyngeal samples capturing only a subset of the overall intrahost viral diversity. The findings highlight that negative tests do not rule out the presence of SARS-CoV-2 reservoirs in immunocompromised patients.
El Moussaoui et al., 31 Aug 2024, Belgium, peer-reviewed, 14 authors.
Contact: melmoussaoui@chuliege.be, kdurkin@uliege.be.
Intrahost evolution leading to distinct lineages in the upper and lower respiratory tracts during SARS-CoV-2 prolonged infection
Virus Evolution, doi:10.1093/ve/veae073
Accumulating evidence points to persistent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections in immunocompromised individuals as a source of novel lineages. While intrahost evolution of the virus in chronically infected patients has previously been reported, existing knowledge is primarily based on samples from the nasopharynx. In this study, we investigate the intrahost evolution and genetic diversity that accumulated during a prolonged SARS-CoV-2 infection with the Omicron BF.7 sublineage, which is estimated to have persisted for >1 year in an immunosuppressed patient. Based on the sequencing of eight samples collected at six time points, we identified 87 intrahost single-nucleotide variants, 2 indels, and a 362-bp deletion. Our analysis revealed distinct viral genotypes in the nasopharyngeal (NP), endotracheal aspirate, and bronchoalveolar lavage samples. This suggests that NP samples may not offer a comprehensive representation of the overall intrahost viral diversity. Our findings not only demonstrate that the Omicron BF.7 sublineage can further diverge from its already exceptionally mutated state but also highlight that patients chronically infected with SARS-CoV-2 can develop genetically specific viral populations across distinct anatomic compartments. This provides novel insights into the intricate nature of viral diversity and evolution dynamics in persistent infections.
Table S4 . Variant Call Format files (VCFs) and BAMs were checked in the Integrative Genomics Viewer (Thorvaldsdóttir et al. 2013) . The consensus sequence for the long-range PCR products was generated using the bcftools consensus command ( https://samtools. github.io/bcftools/bcftools.html ) and manually incorporated into the ARTIC network field bioinformatics pipeline consensus. In order to generate the patient BF.7 ancestral sequence, we used the bcftools isec tool to select SNVs and deletions with an allele frequency of >98% that are common to all patient samples. We also included an SNV found in the BAL and ETA samples, which overlaps with the 362-bp deletion in the NP samples. The resulting VCF file contains 72 SNVs/deletions, and this file was used in combination with the bcftools consensus tool to produce a consensus sequence for the ancestral lineage.
Phylogenetics Consensus genomes were placed on the existing phylogenetic tree via UShER using the phylogenetic tree generated with genomes from GISAID, GeneBank, COVID-19 Genomics UK Consortium, and China National Center for Bioinformation. In the case of the BF.7 samples, we used the genomes identified by UShER to infer a timescaled phylogenetic tree using the software package BEAST v1.10.5 (Suchard et al. 2018) with the following settings: a skygrid demographic model, a relaxed molecular clock with a normal (8 × 10 -4 , 1 × 10 -4 ) prior distribution, and an HKY + G4 + I substitution model (Hasegawa et al...
References
Artesi, Bontems, Göbbels, A recurrent mutation at position 26340 of SARS-CoV-2 is associated with failure of the E gene quantitative reverse transcription-PCR utilized in a commercial dual-target diagnostic assay, J Clin Microbiol
Avanzato, Matson, Seifert, Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer, Cell
Bloom, Neher, Fitness effects of mutations to SARS-CoV-2 proteins, Virus Evol
Bollen, Artesi, Durkin, Exploiting genomic surveillance to map the spatio-temporal dispersal of SARS-CoV-2 spike mutations in Belgium across 2020, Sci Rep
Bussani, Zentilin, Correa, Persistent SARS-CoV-2 infection in patients seemingly recovered from COVID-19, J Pathol
Carabelli, Peacock, Thorne, SARS-CoV-2 variant biology: immune escape, transmission and fitness, Nat Rev Microbiol
Chaguza, Hahn, Petrone, Accelerated SARS-CoV-2 intrahost evolution leading to distinct genotypes during chronic infection, Cell Rep Med
Chen, Chin, Kenney, Spike and nsp6 are key determinants of SARS-CoV-2 Omicron BA.1 attenuation, Nature
Choi, Choudhary, Persistence and evolution of SARS-CoV-2 in an immunocompromised host, New Engl J Med
Cuypers, Dellicour, Hong, Two years of genomic surveillance in Belgium during the SARS-CoV-2 pandemic to attain country-wide coverage and monitor the introduction and spread of emerging variants, Viruses
Dadonaite, Brown, Mcmahon, Spike deep mutational scanning helps predict success of SARS-CoV-2 clades, Nature
Dellicour, Durkin, Hong, A phylodynamic workflow to rapidly gain insights into the dispersal history and dynamics of SARS-CoV-2 lineages, Mol Biol Evol
Faria, Mellan, Whittaker, Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil, Science
Farjo, Koelle, Martin, Within-host evolutionary dynamics and tissue compartmentalization during acute SARS-CoV-2 infection, J Virol
Feng, Bevins, Chandler, Transmission of SARS-CoV-2 in free-ranging white-tailed deer in the United States, Nat Commun
Freed, Vlková, Faisal, Rapid and inexpensive wholegenome sequencing of SARS-CoV-2 using 1200 bp tiled amplicons and Oxford Nanopore Rapid Barcoding, Biol Methods Protoc
Fung, Siu, SARS-CoV-2 main protease suppresses type I interferon production by preventing nuclear translocation of phosphorylated IRF3, Int J Bio Sci
Gong, Tsao, Hsiao, SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East, Emerging Microbes Infect
Gregory, Trujillo, Rushford, Genetic diversity and evolutionary convergence of cryptic SARS-CoV-2 lineages detected via wastewater sequencing, PLoS Pathogens
Guilbaud, Yusti, Leducq, Higher levels of SARS-CoV-2 genetic variation in immunocompromised patients: a retrospective case-control study, J Infect Dis
Harari, Miller, Fleishon, Using big sequencing data to identify chronic SARS-Coronavirus-2 infections, Nat Commun
Hasegawa, Kishino, Yano, Dating of the human-ape splitting by a molecular clock of mitochondrial DNA, J Mol Evol
Hill, Plessis, Peacock, The origins and molecular evolution of SARS-CoV-2 lineage B.1.1.7 in the UK, Virus Evol
Hui, Ho, Cheung, SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo, Nature
Jung, Kmiec, Koepke, Omicron: what makes the latest SARS-CoV-2 variant of concern so concerning?, J Virol
Kaku, Okumura, Padilla-Blanco, Virological characteristics of the SARS-CoV-2 JN.1 variant, Lancet Infect Dis
Kemp, Collier, Datir, SARS-CoV-2 evolution during treatment of chronic infection, Nature
Kim, Lee, Khalid, SARS-CoV-2 protein ORF8 limits expression levels of Spike antigen and facilitates immune evasion of infected host cells, J Biol Chem
Machkovech, Hahn, Wang, Persistent SARS-CoV-2 infection: significance and implications, Lancet Infect Dis
Markov, Ghafari, Beer, The evolution of SARS-CoV-2, Nat Rev Microbiol
Mazur-Panasiuk, Rabalski, Gromowski, Expansion of a SARS-CoV-2 Delta variant with an 872 nt deletion encompassing ORF7a, ORF7b and ORF8, Euro Surveillance
Nabieva, Komissarov, Klink, A highly divergent SARS-CoV-2 lineage B.1.1 sample in a patient with long-term COVID-19, MedRxiv
Obermeyer, Jankowiak, Barkas, Analysis of 6.4 million SARS-CoV-2 genomes identifies mutations associated with fitness, Science
Parsons, Acharya, Evolution of the SARS-CoV-2 Omicron spike, Cell Rep
Patchett, Lv, Rut, A molecular sensor determines the ubiquitin substrate specificity of SARS-CoV-2 papain-like protease, Cell Rep
Pather, Madhi, Cowling, SARS-CoV-2 Omicron variants: burden of disease, impact on vaccine effectiveness and need for variant-adapted vaccines, Front Immunol
Popa, Genger, Nicholson, Genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2, Sci Trans Med
Proal, Vanelzakker, Aleman, SARS-CoV-2 reservoir in post-acute sequelae of COVID-19 (PASC), Nat Immunol
Raglow, Surie, Chappell, SARS-CoV-2 shedding and evolution in patients who were immunocompromised during the omicron period: a multicentre, prospective analysis, Lancet Microbe
Rambaut, Drummond, Xie, Posterior summarization in Bayesian phylogenetics using tracer 1.7, Syst Biol
Roemer, Sheward, Hisner, SARS-CoV-2 evolution in the Omicron era, Nat Microbiol
Sanderson, Hisner, Donovan-Banfield, A molnupiravirassociated mutational signature in global SARS-CoV-2 genomes, Nature
Sonnleitner, Sonnleitner, Hinterbichler, The mutational dynamics of the SARS-CoV-2 virus in serial passages in vitro, Virologica Sin
Stein, Ramelli, Grazioli, SARS-CoV-2 infection and persistence in the human body and brain at autopsy, Nature
Su, Anderson, Be, Discovery and genomic characterization of a 382-nucleotide deletion in ORF7b and ORF8 during the early evolution of SARS-CoV-2, mBio
Suchard, Lemey, Baele, Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10, Virus Evol
Tang, Yu, Guo, Clinical characteristics and host immunity responses of SARS-CoV-2 Omicron variant BA.2 with deletion of ORF7a, ORF7b and ORF8, Virol J
Thorvaldsdóttir, Robinson, Mesirov, Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration, Briefings Bioinf
Tonkin-Hill, Martincorena, Amato, Patterns of within-host genetic diversity in SARS-CoV-2, eLife
Turakhia, Thornlow, Hinrichs, Ultrafast Sample placement on Existing tRees (UShER) enables real-time phylogenetics for the SARS-CoV-2 pandemic, Nat Genet
Van Cleemput, Van Snippenberg, Lambrechts, Organspecific genome diversity of replication-competent SARS-CoV-2, Nat Commun
Van Der Moeren, Selhorst, Ha, Viral evolution and immunology of SARS-CoV-2 in a persistent infection after treatment with rituximab, Viruses
Viana, Moyo, Amoako, Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa, Nature
Wagner, Kistler, Perchetti, Positive selection underlies repeated knockout of ORF8 in SARS-CoV-2 evolution, Nat Commun
Wilkinson, Richter, Casey, Recurrent SARS-CoV-2 mutations in immunodeficient patients, Virus Evol
Wilm, Aw, Bertrand, LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets, Nucleic Acids Res
Yang, Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods, J Mol Evol
Yi, Wang, Wang, The emergence and spread of novel SARS-CoV-2 variants, Front Public Health
Young, Fong, Chan, Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study, Lancet
Zheng, Li, Su, Symphonizing pileup and full-alignment for deep learning-based long-read variant calling, Nat Comput Sci
Zinzula, Lost in deletion: the enigmatic ORF8 protein of SARS-CoV-2, Biochem Biophys Res Commun
DOI record:
{
"DOI": "10.1093/ve/veae073",
"ISSN": [
"2057-1577"
],
"URL": "http://dx.doi.org/10.1093/ve/veae073",
"abstract": "<jats:title>Abstract</jats:title>\n <jats:p>Accumulating evidence points to persistent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections in immunocompromised individuals as a source of novel lineages. While intrahost evolution of the virus in chronically infected patients has previously been reported, existing knowledge is primarily based on samples from the nasopharynx. In this study, we investigate the intrahost evolution and genetic diversity that accumulated during a prolonged SARS-CoV-2 infection with the Omicron BF.7 sublineage, which is estimated to have persisted for &gt;1 year in an immunosuppressed patient. Based on the sequencing of eight samples collected at six time points, we identified 87 intrahost single-nucleotide variants, 2 indels, and a 362-bp deletion. Our analysis revealed distinct viral genotypes in the nasopharyngeal (NP), endotracheal aspirate, and bronchoalveolar lavage samples. This suggests that NP samples may not offer a comprehensive representation of the overall intrahost viral diversity. Our findings not only demonstrate that the Omicron BF.7 sublineage can further diverge from its already exceptionally mutated state but also highlight that patients chronically infected with SARS-CoV-2 can develop genetically specific viral populations across distinct anatomic compartments. This provides novel insights into the intricate nature of viral diversity and evolution dynamics in persistent infections.</jats:p>",
"author": [
{
"affiliation": [
{
"name": "Department of Infectious Diseases and General Internal Medicine, University Hospital of Liège , 1 Avenue de l’Hôpital, Liège 4000,",
"place": [
"Belgium"
]
}
],
"family": "El Moussaoui",
"given": "Majdouline",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Department of Microbiology, University Hospital of Liège , 1 Avenue de l’Hôpital, Liège 4000,",
"place": [
"Belgium"
]
}
],
"family": "Bontems",
"given": "Sebastien",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Microbiology, University Hospital of Liège , 1 Avenue de l’Hôpital, Liège 4000,",
"place": [
"Belgium"
]
}
],
"family": "Meex",
"given": "Cecile",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Microbiology, University Hospital of Liège , 1 Avenue de l’Hôpital, Liège 4000,",
"place": [
"Belgium"
]
}
],
"family": "Hayette",
"given": "Marie-Pierre",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Hematology, University Hospital of Liège , 1 Avenue de l’Hôpital, Liège 4000,",
"place": [
"Belgium"
]
}
],
"family": "Lejeune",
"given": "Marie",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-6354-4943",
"affiliation": [
{
"name": "Department of Microbiology, Immunology and Transplantation, Laboratory for Clinical and Epidemiological Virology, Rega Institute, KU Leuven , 49 Herestraat, Leuven 3000,",
"place": [
"Belgium"
]
}
],
"authenticated-orcid": false,
"family": "Hong",
"given": "Samuel L",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-9558-1052",
"affiliation": [
{
"name": "Department of Microbiology, Immunology and Transplantation, Laboratory for Clinical and Epidemiological Virology, Rega Institute, KU Leuven , 49 Herestraat, Leuven 3000,",
"place": [
"Belgium"
]
},
{
"name": "Spatial Epidemiology Lab (SpELL), Université Libre de Bruxelles , 50 Avenue Franklin Roosevelt, Bruxelles 1050,",
"place": [
"Belgium"
]
}
],
"authenticated-orcid": false,
"family": "Dellicour",
"given": "Simon",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Infectious Diseases and General Internal Medicine, University Hospital of Liège , 1 Avenue de l’Hôpital, Liège 4000,",
"place": [
"Belgium"
]
}
],
"family": "Moutschen",
"given": "Michel",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Human Genetics, University Hospital of Liège , 1 Avenue de l’Hôpital, Liège 4000,",
"place": [
"Belgium"
]
},
{
"name": "Laboratory of Human Genetics, GIGA Institute, University of Liège , 1 Avenue de l’Hôpital, Liège 4000,",
"place": [
"Belgium"
]
}
],
"family": "Cambisano",
"given": "Nadine",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Human Genetics, University Hospital of Liège , 1 Avenue de l’Hôpital, Liège 4000,",
"place": [
"Belgium"
]
},
{
"name": "Laboratory of Human Genetics, GIGA Institute, University of Liège , 1 Avenue de l’Hôpital, Liège 4000,",
"place": [
"Belgium"
]
}
],
"family": "Renotte",
"given": "Nathalie",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Human Genetics, University Hospital of Liège , 1 Avenue de l’Hôpital, Liège 4000,",
"place": [
"Belgium"
]
},
{
"name": "Laboratory of Human Genetics, GIGA Institute, University of Liège , 1 Avenue de l’Hôpital, Liège 4000,",
"place": [
"Belgium"
]
}
],
"family": "Bours",
"given": "Vincent",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-8192-1351",
"affiliation": [
{
"name": "Department of Infectious Diseases and General Internal Medicine, University Hospital of Liège , 1 Avenue de l’Hôpital, Liège 4000,",
"place": [
"Belgium"
]
}
],
"authenticated-orcid": false,
"family": "Darcis",
"given": "Gilles",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Human Genetics, University Hospital of Liège , 1 Avenue de l’Hôpital, Liège 4000,",
"place": [
"Belgium"
]
},
{
"name": "Laboratory of Human Genetics, GIGA Institute, University of Liège , 1 Avenue de l’Hôpital, Liège 4000,",
"place": [
"Belgium"
]
}
],
"family": "Artesi",
"given": "Maria",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-7475-6618",
"affiliation": [
{
"name": "Department of Human Genetics, University Hospital of Liège , 1 Avenue de l’Hôpital, Liège 4000,",
"place": [
"Belgium"
]
},
{
"name": "Laboratory of Human Genetics, GIGA Institute, University of Liège , 1 Avenue de l’Hôpital, Liège 4000,",
"place": [
"Belgium"
]
}
],
"authenticated-orcid": false,
"family": "Durkin",
"given": "Keith",
"sequence": "additional"
}
],
"container-title": "Virus Evolution",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
8,
30
]
],
"date-time": "2024-08-30T00:09:16Z",
"timestamp": 1724976556000
},
"deposited": {
"date-parts": [
[
2024,
10,
12
]
],
"date-time": "2024-10-12T15:37:07Z",
"timestamp": 1728747427000
},
"funder": [
{
"award": [
"101094685"
],
"name": "LEAPS"
},
{
"DOI": "10.13039/100010661",
"award": [
"874850"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100010661",
"id-type": "DOI"
}
],
"name": "Horizon 2020 Framework Programme"
},
{
"award": [
"1710180"
],
"name": "Région Wallone"
},
{
"award": [
"F.4515.22 H.C.008.20"
],
"name": "Fonds De La Recherche Scientifique – FNRS"
},
{
"DOI": "10.13039/501100003130",
"award": [
"G098321N"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100003130",
"id-type": "DOI"
}
],
"name": "Fonds Wetenschappelijk Onderzoek"
},
{
"award": [
"101094685"
],
"name": "LEAPS"
},
{
"DOI": "10.13039/100010661",
"award": [
"874850"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100010661",
"id-type": "DOI"
}
],
"name": "Horizon 2020 Framework Programme"
},
{
"award": [
"1710180"
],
"name": "Région Wallone"
},
{
"award": [
"F.4515.22 H.C.008.20"
],
"name": "Fonds De La Recherche Scientifique – FNRS"
},
{
"DOI": "10.13039/501100003130",
"award": [
"G098321N"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100003130",
"id-type": "DOI"
}
],
"name": "Fonds Wetenschappelijk Onderzoek"
}
],
"indexed": {
"date-parts": [
[
2024,
10,
12
]
],
"date-time": "2024-10-12T16:10:12Z",
"timestamp": 1728749412742
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2024
]
]
},
"journal-issue": {
"issue": "1",
"published-print": {
"date-parts": [
[
2024,
10,
12
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 243,
"start": {
"date-parts": [
[
2024,
8,
31
]
],
"date-time": "2024-08-31T00:00:00Z",
"timestamp": 1725062400000
}
}
],
"link": [
{
"URL": "https://academic.oup.com/ve/advance-article-pdf/doi/10.1093/ve/veae073/58985308/veae073.pdf",
"content-type": "application/pdf",
"content-version": "am",
"intended-application": "syndication"
},
{
"URL": "https://academic.oup.com/ve/article-pdf/10/1/veae073/59735202/veae073.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "syndication"
},
{
"URL": "https://academic.oup.com/ve/article-pdf/10/1/veae073/59735202/veae073.pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "286",
"original-title": [],
"prefix": "10.1093",
"published": {
"date-parts": [
[
2024
]
]
},
"published-online": {
"date-parts": [
[
2024,
8,
31
]
]
},
"published-other": {
"date-parts": [
[
2024
]
]
},
"published-print": {
"date-parts": [
[
2024,
10,
12
]
]
},
"publisher": "Oxford University Press (OUP)",
"reference": [
{
"DOI": "10.1128/JCM.01598-20",
"article-title": "A recurrent mutation at position 26340 of SARS-CoV-2 is associated with failure of the E gene quantitative reverse transcription-PCR utilized in a commercial dual-target diagnostic assay",
"author": "Artesi",
"doi-asserted-by": "publisher",
"first-page": "10",
"journal-title": "J Clin Microbiol",
"key": "2024101215362809400_R1",
"volume": "58",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.10.049",
"article-title": "Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer",
"author": "Avanzato",
"doi-asserted-by": "publisher",
"first-page": "1901",
"journal-title": "Cell",
"key": "2024101215362809400_R2",
"volume": "183",
"year": "2020"
},
{
"DOI": "10.1093/ve/vead055",
"article-title": "Fitness effects of mutations to SARS-CoV-2 proteins",
"author": "Bloom",
"doi-asserted-by": "publisher",
"journal-title": "Virus Evol",
"key": "2024101215362809400_R3",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.1038/s41598-021-97667-9",
"article-title": "Exploiting genomic surveillance to map the spatio-temporal dispersal of SARS-CoV-2 spike mutations in Belgium across 2020",
"author": "Bollen",
"doi-asserted-by": "publisher",
"journal-title": "Sci Rep",
"key": "2024101215362809400_R4",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.1002/path.6035",
"article-title": "Persistent SARS-CoV-2 infection in patients seemingly recovered from COVID-19",
"author": "Bussani",
"doi-asserted-by": "publisher",
"first-page": "254",
"journal-title": "J Pathol",
"key": "2024101215362809400_R5",
"volume": "259",
"year": "2023"
},
{
"DOI": "10.1038/s41579-022-00841-7",
"article-title": "SARS-CoV-2 variant biology: immune escape, transmission and fitness",
"author": "Carabelli",
"doi-asserted-by": "publisher",
"first-page": "162",
"journal-title": "Nat Rev Microbiol",
"key": "2024101215362809400_R6",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.1016/j.xcrm.2023.100943",
"article-title": "Accelerated SARS-CoV-2 intrahost evolution leading to distinct genotypes during chronic infection",
"author": "Chaguza",
"doi-asserted-by": "publisher",
"journal-title": "Cell Rep Med",
"key": "2024101215362809400_R7",
"volume": "4",
"year": "2023"
},
{
"DOI": "10.1038/s41586-023-05697-2",
"article-title": "Spike and nsp6 are key determinants of SARS-CoV-2 Omicron BA.1 attenuation",
"author": "Chen",
"doi-asserted-by": "publisher",
"first-page": "143",
"journal-title": "Nature",
"key": "2024101215362809400_R8",
"volume": "615",
"year": "2023"
},
{
"DOI": "10.1056/NEJMc2031364",
"article-title": "Persistence and evolution of SARS-CoV-2 in an immunocompromised host",
"author": "Choi",
"doi-asserted-by": "publisher",
"first-page": "2291",
"journal-title": "New Engl J Med",
"key": "2024101215362809400_R9",
"volume": "383",
"year": "2020"
},
{
"DOI": "10.3390/v14102301",
"article-title": "Two years of genomic surveillance in Belgium during the SARS-CoV-2 pandemic to attain country-wide coverage and monitor the introduction and spread of emerging variants",
"author": "Cuypers",
"doi-asserted-by": "publisher",
"journal-title": "Viruses",
"key": "2024101215362809400_R10",
"volume": "14",
"year": "2022"
},
{
"DOI": "10.1038/s41586-024-07636-1",
"article-title": "Spike deep mutational scanning helps predict success of SARS-CoV-2 clades",
"author": "Dadonaite",
"doi-asserted-by": "publisher",
"first-page": "617",
"journal-title": "Nature",
"key": "2024101215362809400_R11",
"volume": "631",
"year": "2024"
},
{
"DOI": "10.1093/molbev/msaa284",
"article-title": "A phylodynamic workflow to rapidly gain insights into the dispersal history and dynamics of SARS-CoV-2 lineages",
"author": "Dellicour",
"doi-asserted-by": "publisher",
"first-page": "1608",
"journal-title": "Mol Biol Evol",
"key": "2024101215362809400_R12",
"volume": "38",
"year": "2021"
},
{
"DOI": "10.1126/science.abh2644",
"article-title": "Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil",
"author": "Faria",
"doi-asserted-by": "publisher",
"first-page": "815",
"journal-title": "Science",
"key": "2024101215362809400_R13",
"volume": "372",
"year": "2021"
},
{
"DOI": "10.1128/jvi.01618-23",
"article-title": "Within-host evolutionary dynamics and tissue compartmentalization during acute SARS-CoV-2 infection",
"author": "Farjo",
"doi-asserted-by": "publisher",
"journal-title": "J Virol",
"key": "2024101215362809400_R14",
"volume": "98",
"year": "2024"
},
{
"DOI": "10.1038/s41467-023-39782-x",
"article-title": "Transmission of SARS-CoV-2 in free-ranging white-tailed deer in the United States",
"author": "Feng",
"doi-asserted-by": "publisher",
"journal-title": "Nat Commun",
"key": "2024101215362809400_R15",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1093/biomethods/bpaa014",
"article-title": "Rapid and inexpensive whole-genome sequencing of SARS-CoV-2 using 1200 bp tiled amplicons and Oxford Nanopore Rapid Barcoding",
"author": "Freed",
"doi-asserted-by": "publisher",
"journal-title": "Biol Methods Protoc",
"key": "2024101215362809400_R16",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.7150/ijbs.59943",
"article-title": "SARS-CoV-2 main protease suppresses type I interferon production by preventing nuclear translocation of phosphorylated IRF3",
"author": "Fung",
"doi-asserted-by": "publisher",
"first-page": "1547",
"journal-title": "Int J Bio Sci",
"key": "2024101215362809400_R17",
"volume": "17",
"year": "2021"
},
{
"DOI": "10.1080/22221751.2020.1782271",
"article-title": "SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East",
"author": "Gong",
"doi-asserted-by": "publisher",
"first-page": "1457",
"journal-title": "Emerging Microbes Infect",
"key": "2024101215362809400_R18",
"volume": "9",
"year": "2020"
},
{
"DOI": "10.1371/journal.ppat.1010636",
"article-title": "Genetic diversity and evolutionary convergence of cryptic SARS- CoV-2 lineages detected via wastewater sequencing",
"author": "Gregory",
"doi-asserted-by": "publisher",
"journal-title": "PLoS Pathogens",
"key": "2024101215362809400_R19",
"volume": "18",
"year": "2022"
},
{
"DOI": "10.1093/infdis/jiad499",
"article-title": "Higher levels of SARS-CoV-2 genetic variation in immunocompromised patients: a retrospective case-control study",
"author": "Guilbaud",
"doi-asserted-by": "publisher",
"first-page": "1041",
"journal-title": "J Infect Dis",
"key": "2024101215362809400_R20",
"volume": "229",
"year": "2024"
},
{
"DOI": "10.1038/s41467-024-44803-4",
"article-title": "Using big sequencing data to identify chronic SARS-Coronavirus-2 infections",
"author": "Harari",
"doi-asserted-by": "publisher",
"journal-title": "Nat Commun",
"key": "2024101215362809400_R21",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1007/bf02101694",
"article-title": "Dating of the human-ape splitting by a molecular clock of mitochondrial DNA",
"author": "Hasegawa",
"doi-asserted-by": "publisher",
"first-page": "160",
"journal-title": "J Mol Evol",
"key": "2024101215362809400_R22",
"volume": "22",
"year": "1985"
},
{
"DOI": "10.1093/ve/veac080",
"article-title": "The origins and molecular evolution of SARS-CoV-2 lineage B.1.1.7 in the UK",
"author": "Hill",
"doi-asserted-by": "publisher",
"journal-title": "Virus Evol",
"key": "2024101215362809400_R23",
"volume": "8",
"year": "2022"
},
{
"DOI": "10.1038/s41586-022-04479-6",
"article-title": "SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo",
"author": "Hui",
"doi-asserted-by": "publisher",
"first-page": "715",
"journal-title": "Nature",
"key": "2024101215362809400_R24",
"volume": "603",
"year": "2022"
},
{
"DOI": "10.1128/jvi.02077-21",
"article-title": "Omicron: what makes the latest SARS-CoV-2 variant of concern so concerning?",
"author": "Jung",
"doi-asserted-by": "publisher",
"journal-title": "J Virol",
"key": "2024101215362809400_R25",
"volume": "96",
"year": "2022"
},
{
"DOI": "10.1016/S1473-3099(23)00813-7",
"article-title": "Virological characteristics of the SARS-CoV-2 JN.1 variant",
"author": "Kaku",
"doi-asserted-by": "publisher",
"journal-title": "Lancet Infect Dis",
"key": "2024101215362809400_R26",
"volume": "24",
"year": "2024"
},
{
"DOI": "10.1038/s41586-021-03291-y",
"article-title": "SARS-CoV-2 evolution during treatment of chronic infection",
"author": "Kemp",
"doi-asserted-by": "publisher",
"first-page": "277",
"journal-title": "Nature",
"key": "2024101215362809400_R27",
"volume": "592",
"year": "2021"
},
{
"DOI": "10.1016/j.jbc.2023.104955",
"article-title": "SARS-CoV-2 protein ORF8 limits expression levels of Spike antigen and facilitates immune evasion of infected host cells",
"author": "Kim",
"doi-asserted-by": "publisher",
"journal-title": "J Biol Chem",
"key": "2024101215362809400_R28",
"volume": "299",
"year": "2023"
},
{
"DOI": "10.1016/S1473-3099(23)00815-0",
"article-title": "Persistent SARS-CoV-2 infection: significance and implications",
"author": "Machkovech",
"doi-asserted-by": "publisher",
"first-page": "e453",
"journal-title": "Lancet Infect Dis",
"key": "2024101215362809400_R29",
"volume": "24",
"year": "2024"
},
{
"DOI": "10.1038/s41579-023-00878-2",
"article-title": "The evolution of SARS-CoV-2",
"author": "Markov",
"doi-asserted-by": "publisher",
"first-page": "361",
"journal-title": "Nat Rev Microbiol",
"key": "2024101215362809400_R30",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.2807/1560-7917.ES.2021.26.39.2100902",
"article-title": "Expansion of a SARS-CoV-2 Delta variant with an 872 nt deletion encompassing ORF7a, ORF7b and ORF8, Poland, July to August 2021",
"author": "Mazur-Panasiuk",
"doi-asserted-by": "publisher",
"journal-title": "Euro Surveillance",
"key": "2024101215362809400_R31",
"volume": "26",
"year": "2021"
},
{
"DOI": "10.1101/2023.09.14.23295379",
"article-title": "A highly divergent SARS-CoV-2 lineage B.1.1 sample in a patient with long-term COVID-19",
"author": "Nabieva",
"doi-asserted-by": "publisher",
"journal-title": "MedRxiv",
"key": "2024101215362809400_R32",
"year": "2023"
},
{
"DOI": "10.1126/science.abm1208",
"article-title": "Analysis of 6.4 million SARS-CoV-2 genomes identifies mutations associated with fitness",
"author": "Obermeyer",
"doi-asserted-by": "publisher",
"first-page": "1327",
"journal-title": "Science",
"key": "2024101215362809400_R33",
"volume": "376",
"year": "2022"
},
{
"DOI": "10.1016/j.celrep.2023.113444",
"article-title": "Evolution of the SARS-CoV-2 Omicron spike",
"author": "Parsons",
"doi-asserted-by": "publisher",
"journal-title": "Cell Rep",
"key": "2024101215362809400_R34",
"volume": "42",
"year": "2023"
},
{
"DOI": "10.1016/j.celrep.2021.109754",
"article-title": "A molecular sensor determines the ubiquitin substrate specificity of SARS-CoV-2 papain-like protease",
"author": "Patchett",
"doi-asserted-by": "publisher",
"journal-title": "Cell Rep",
"key": "2024101215362809400_R35",
"volume": "36",
"year": "2021"
},
{
"DOI": "10.3389/fimmu.2023.1130539",
"article-title": "SARS-CoV-2 Omicron variants: burden of disease, impact on vaccine effectiveness and need for variant-adapted vaccines",
"author": "Pather",
"doi-asserted-by": "publisher",
"journal-title": "Front Immunol",
"key": "2024101215362809400_R36",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1126/scitranslmed.abe2555",
"article-title": "Genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2",
"author": "Popa",
"doi-asserted-by": "publisher",
"journal-title": "Sci Trans Med",
"key": "2024101215362809400_R37",
"volume": "12",
"year": "2020"
},
{
"DOI": "10.1038/s41590-023-01601-2",
"article-title": "SARS-CoV-2 reservoir in post-acute sequelae of COVID-19 (PASC)",
"author": "Proal",
"doi-asserted-by": "publisher",
"first-page": "1616",
"journal-title": "Nat Immunol",
"key": "2024101215362809400_R38",
"volume": "24",
"year": "2023"
},
{
"DOI": "10.1016/S2666-5247(23)00336-1",
"article-title": "SARS-CoV-2 shedding and evolution in patients who were immunocompromised during the omicron period: a multicentre, prospective analysis",
"author": "Raglow",
"doi-asserted-by": "publisher",
"first-page": "e235",
"journal-title": "Lancet Microbe",
"key": "2024101215362809400_R39",
"volume": "5",
"year": "2024"
},
{
"DOI": "10.1093/sysbio/syy032",
"article-title": "Posterior summarization in Bayesian phylogenetics using tracer 1.7",
"author": "Rambaut",
"doi-asserted-by": "publisher",
"first-page": "901",
"journal-title": "Syst Biol",
"key": "2024101215362809400_R40",
"volume": "67",
"year": "2018"
},
{
"DOI": "10.1038/s41564-023-01504-w",
"article-title": "SARS-CoV-2 evolution in the Omicron era",
"author": "Roemer",
"doi-asserted-by": "publisher",
"first-page": "1952",
"journal-title": "Nat Microbiol",
"key": "2024101215362809400_R41",
"volume": "8",
"year": "2023"
},
{
"DOI": "10.1038/s41586-023-06649-6",
"article-title": "A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes",
"author": "Sanderson",
"doi-asserted-by": "publisher",
"first-page": "594",
"journal-title": "Nature",
"key": "2024101215362809400_R42",
"volume": "623",
"year": "2023"
},
{
"DOI": "10.1016/j.virs.2022.01.029",
"article-title": "The mutational dynamics of the SARS-CoV-2 virus in serial passages in vitro",
"author": "Sonnleitner",
"doi-asserted-by": "publisher",
"first-page": "198",
"journal-title": "Virologica Sin",
"key": "2024101215362809400_R43",
"volume": "37",
"year": "2022"
},
{
"DOI": "10.1038/s41586-022-05542-y",
"article-title": "SARS-CoV-2 infection and persistence in the human body and brain at autopsy",
"author": "Stein",
"doi-asserted-by": "publisher",
"first-page": "758",
"journal-title": "Nature",
"key": "2024101215362809400_R44",
"volume": "612",
"year": "2022"
},
{
"DOI": "10.1128/mBio.01610-20",
"article-title": "Discovery and genomic characterization of a 382-nucleotide deletion in ORF7b and ORF8 during the early evolution of SARS-CoV-2",
"author": "Su",
"doi-asserted-by": "publisher",
"first-page": "10",
"journal-title": "mBio",
"key": "2024101215362809400_R45",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1093/ve/vey016",
"article-title": "Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10",
"author": "Suchard",
"doi-asserted-by": "publisher",
"journal-title": "Virus Evol",
"key": "2024101215362809400_R46",
"volume": "4",
"year": "2018"
},
{
"DOI": "10.1186/s12985-023-02066-3",
"article-title": "Clinical characteristics and host immunity responses of SARS-CoV-2 Omicron variant BA.2 with deletion of ORF7a, ORF7b and ORF8",
"author": "Tang",
"doi-asserted-by": "publisher",
"journal-title": "Virol J",
"key": "2024101215362809400_R47",
"volume": "20",
"year": "2023"
},
{
"DOI": "10.1093/bib/bbs017",
"article-title": "Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration",
"author": "Thorvaldsdóttir",
"doi-asserted-by": "publisher",
"first-page": "178",
"journal-title": "Briefings Bioinf",
"key": "2024101215362809400_R48",
"volume": "14",
"year": "2013"
},
{
"DOI": "10.7554/eLife.66857",
"article-title": "Patterns of within-host genetic diversity in SARS-CoV-2",
"author": "Tonkin-Hill",
"doi-asserted-by": "publisher",
"journal-title": "eLife",
"key": "2024101215362809400_R49",
"volume": "10",
"year": "2021"
},
{
"DOI": "10.1038/s41588-021-00862-7",
"article-title": "Ultrafast Sample placement on Existing tRees (UShER) enables real-time phylogenetics for the SARS-CoV-2 pandemic",
"author": "Turakhia",
"doi-asserted-by": "publisher",
"first-page": "809",
"journal-title": "Nat Genet",
"key": "2024101215362809400_R50",
"volume": "53",
"year": "2021"
},
{
"DOI": "10.1038/s41467-021-26884-7",
"article-title": "Organ-specific genome diversity of replication-competent SARS-CoV-2",
"author": "Van Cleemput",
"doi-asserted-by": "publisher",
"journal-title": "Nat Commun",
"key": "2024101215362809400_R51",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.3390/v14040752",
"article-title": "Viral evolution and immunology of SARS-CoV-2 in a persistent infection after treatment with rituximab",
"author": "Van der Moeren",
"doi-asserted-by": "publisher",
"journal-title": "Viruses",
"key": "2024101215362809400_R52",
"volume": "14",
"year": "2022"
},
{
"DOI": "10.1038/s41586-022-04411-y",
"article-title": "Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa",
"author": "Viana",
"doi-asserted-by": "publisher",
"first-page": "679",
"journal-title": "Nature",
"key": "2024101215362809400_R53",
"volume": "603",
"year": "2022"
},
{
"DOI": "10.1038/s41467-024-47599-5",
"article-title": "Positive selection underlies repeated knockout of ORF8 in SARS-CoV-2 evolution",
"author": "Wagner",
"doi-asserted-by": "publisher",
"journal-title": "Nat Commun",
"key": "2024101215362809400_R54",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1093/ve/veac050",
"article-title": "Recurrent SARS-CoV-2 mutations in immunodeficient patients",
"author": "Wilkinson",
"doi-asserted-by": "publisher",
"journal-title": "Virus Evol",
"key": "2024101215362809400_R55",
"volume": "8",
"year": "2022"
},
{
"DOI": "10.1093/nar/gks918",
"article-title": "LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets",
"author": "Wilm",
"doi-asserted-by": "publisher",
"first-page": "11189",
"journal-title": "Nucleic Acids Res",
"key": "2024101215362809400_R56",
"volume": "40",
"year": "2012"
},
{
"DOI": "10.1007/bf00160154",
"article-title": "Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods",
"author": "Yang",
"doi-asserted-by": "publisher",
"first-page": "306",
"journal-title": "J Mol Evol",
"key": "2024101215362809400_R57",
"volume": "39",
"year": "1994"
},
{
"DOI": "10.3389/fpubh.2021.696664",
"article-title": "The emergence and spread of novel SARS-CoV-2 variants",
"author": "Yi",
"doi-asserted-by": "publisher",
"journal-title": "Front Public Health",
"key": "2024101215362809400_R58",
"volume": "9",
"year": "2021"
},
{
"DOI": "10.1016/S0140-6736(20)31757-8",
"article-title": "Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study",
"author": "Young",
"doi-asserted-by": "publisher",
"first-page": "603",
"journal-title": "Lancet",
"key": "2024101215362809400_R59",
"volume": "396",
"year": "2020"
},
{
"DOI": "10.1038/s43588-022-00387-x",
"article-title": "Symphonizing pileup and full-alignment for deep learning-based long-read variant calling",
"author": "Zheng",
"doi-asserted-by": "publisher",
"first-page": "797",
"journal-title": "Nat Comput Sci",
"key": "2024101215362809400_R60",
"volume": "2",
"year": "2022"
},
{
"DOI": "10.1016/j.bbrc.2020.10.045",
"article-title": "Lost in deletion: the enigmatic ORF8 protein of SARS-CoV-2",
"author": "Zinzula",
"doi-asserted-by": "publisher",
"first-page": "116",
"journal-title": "Biochem Biophys Res Commun",
"key": "2024101215362809400_R61",
"volume": "538",
"year": "2021"
}
],
"reference-count": 61,
"references-count": 61,
"relation": {},
"resource": {
"primary": {
"URL": "https://academic.oup.com/ve/article/doi/10.1093/ve/veae073/7746671"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Intrahost evolution leading to distinct lineages in the upper and lower respiratory tracts during SARS-CoV-2 prolonged infection",
"type": "journal-article",
"volume": "10"
}