Identification and targeting of regulators of SARS-CoV-2–host interactions in the airway epithelium
et al., Science Advances, doi:10.1126/sciadv.adu2079, May 2025
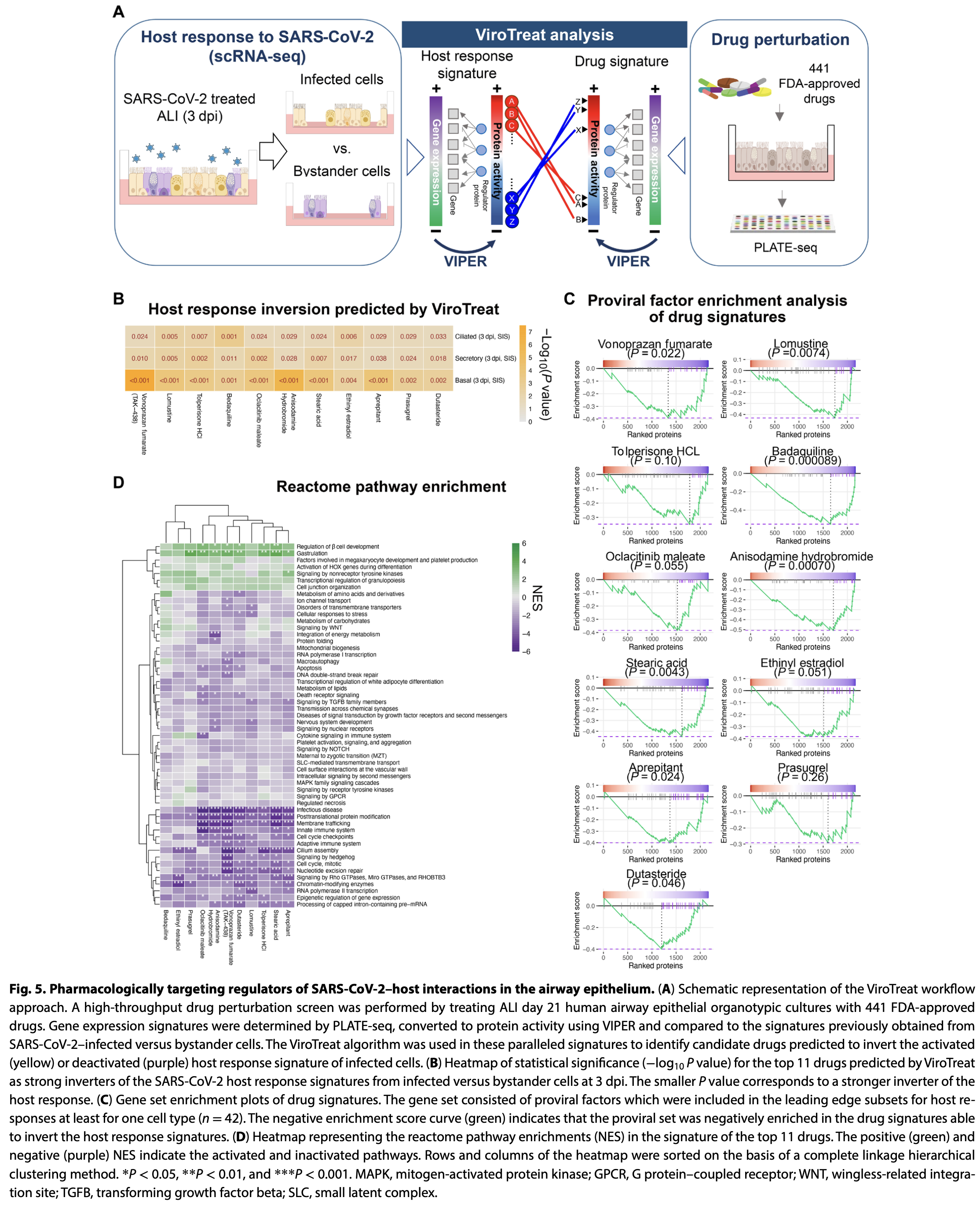
In vitro study of SARS-CoV-2-infected human airway epithelial cells using network-based analysis to identify master regulator proteins controlling viral-host interactions and potential drug candidates. Authors analyzed single-cell transcriptomic profiles from primary human airway epithelial cells grown in organotypic cultures, identifying cell type-specific master regulators in ciliated, secretory, and basal cells. The study revealed chromatin remodeling, endosomal sorting, and ubiquitin pathways as components of viral-host response. Large-scale drug perturbation screens identified 11 FDA-approved drugs capable of inverting the master regulator signatures activated by SARS-CoV-2, including bedaquiline, aprepitant, and dutasteride.
Dirvin et al., 16 May 2025, USA, peer-reviewed, 13 authors.
Contact: ac2248@cumc.columbia.edu.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Identification and targeting of regulators of SARS-CoV-2-host interactions in the airway epithelium
The impact of SARS-CoV-2 in the lung has been extensively studied, yet the molecular regulators of host-cell programs hijacked by the virus in distinct human airway epithelial cell populations remain poorly understood. Some of the reasons include overreliance on transcriptomic profiling and use of nonprimary cell systems. Here we report a network-based analysis of single-cell transcriptomic profiles able to identify master regulator (MR) proteins controlling SARS-CoV-2-mediated reprogramming in pathophysiologically relevant human ciliated, secretory, and basal cells. This underscored chromatin remodeling, endosomal sorting, ubiquitin pathways, as well as proviral factors identified by CRISPR assays as components of the viral-host response in these cells. Large-scale drug perturbation screens revealed 11 candidate drugs able to invert the entire MR signature activated by SARS-CoV-2. Leveraging MR analysis and perturbational profiles of human primary cells represents an innovative approach to investigate pathogen-host interactions in multiple airway conditions for drug prioritization.
Acknowledgments: M.S. and A.G.-S.want to thank R. Albrecht for management and organization of the BSl-3 facility. Funding: this work was supported by the grants nih-nhlBi R35-hl135834 to W.v.c.; nih-nci R35 cA197745, S10 OD012351, S10 OD021764, S10 OD032433, and U01 cA 272610 to A.c.; nih/niAiD R01Ai160706, nih/niDDK R01DK130425 and U19Ai135972 to A.G.-S. Author contributions: W.v.c.: Writing-original draft, conceptualization, investigation, writing-review and editing, methodology, resources, validation, supervision, project administration, and supervision. A.c.: Writing-original draft, conceptualization, writing-review and editing, methodology, resources, funding acquisition, validation, supervision, and project administration. A.G.-S.: conceptualization, writing-review and editing, methodology, resources, funding acquisition, and supervision. B.D.: Writingoriginal draft, conceptualization, investigation, writing-review and editing, methodology, resources, validation, supervision, project administration, and visualization. h.n.: Writingoriginal draft, conceptualization, investigation, writing-review and editing, methodology, data curation, validation, formal analysis, software, and visualization. l.t.: Writing-original draft, conceptualization, investigation, writing-review and editing, methodology, data curation, validation, formal analysis, software, and visualization. D.c.: conceptualization, investigation, writing-review and editing, methodology, data curation,..
References
-K. Chan, Kyba, What is a master regulator?, J. Stem Cell Res. Ther
-R. Hsu, Hubbell-Engler, Adelmant, Huang, Joyce et al., PRMt1mediated translation regulation is a crucial vulnerability of cancer, Cancer Res
Alvarez, Ré, Politi, Lotti, Phani et al., the regulatory machinery of neurodegeneration in in vitro models of amyotrophic lateral sclerosis, Cell Rep
Alvarez, Shen, Giorgi, Lachmann, Ding et al., Functional characterization of somatic mutations in cancer using network-based inference of protein activity, Nat. Genet
Alvarez, Subramaniam, Tang, Grunn, Aburi et al., A precision oncology approach to the pharmacological targeting of mechanistic dependencies in neuroendocrine tumors, Nat. Genet
Aminpour, Tuszynski, how cOviD-19 hijacks the cytoskeleton: therapeutic implications, Life (Basel)
Aran, Wu, Fong, Hsu, Chak et al., Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage, Nat. Immunol
Arriaga, De Almeida, Zou, Douglass, Picech et al., Oncoloop: A network-based precision cancer medicine framework, Cancer Discov
Arumugam, Shin, Schiavone, Tu, Kesner et al., the master regulator protein BAZ2B can reprogram human hematopoietic lineage-committed progenitors into a multipotent State, Cell Rep
Aytes, Mitrofanova, Alvarez, Castillo-Martin, Zheng et al., cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and cenPF that drives prostate cancer malignancy, Cancer Cell
Baggen, Jansen, Daelemans, cellular host factors for SARS-cov-2 infection, Nat. Microbiol
Biering, Sarnik, Wang, Zengel, Leist et al., Genome-wide bidirectional cRiSPR screens identify mucins as host factors modulating SARS-cov-2 infection, Nat. Genet
Birger, Ghandi, Mesirov, Tamayo, the Molecular Signatures Database (MSigDB) hallmark gene set collection, Cell Syst
Bray, Pimentel, Melsted, Pachter, near-optimal probabilistic RnA-seq quantification, Nat. Biotechnol
Brichta, Shin, Jackson-Lewis, Blesa, Yap et al., identification of neurodegenerative factors using translatome-regulatory network analysis, Nat. Neurosci
Bush, Ray, Alvarez, Realubit, Karan et al., PlAte-seq for genome-wide regulatory network analysis of high-throughput screens, Nat. Commun
Carro, Lim, Alvarez, Bollo, Zhao et al., the transcriptional network for mesenchymal transformation of brain tumours, Nature
Castaneda, Jangra, Yurieva, Martinek, Callender et al., Spatiotemporally organized immunomodulatory response to SARS-cov-2 virus in primary human broncho-alveolar epithelia, iScience
Daniloski, Jordan, Wessels, Hoagland, Kasela et al., identification of required host factors for SARS-cov-2 infection in human cells, Cell
Diacon, Pym, Grobusch, Patientia, Rustomjee et al., the diarylquinoline tMc207 for multidrug-resistant tuberculosis, N. Engl. J. Med
Ding, Douglass, Sonabend, Mela, Bose et al., Quantitative assessment of protein activity in orphan tissues and single cells using the metaviPeR algorithm, Nat. Commun
Dong, Zhang, Kuang, Wang, Selective regulation in ribosome biogenesis and protein production for efficient viral translation, Arch. Microbiol
Giorgi, Lopez, Califano, ARAcne-AP: Gene network reverse engineering through adaptive partitioning inference of mutual information, Bioinformatics
Ha, Wang, Rangel, Van Krieken, Liu et al., targeting GRP78 suppresses oncogenic KRAS protein expression and reduces viability of cancer cells bearing various KRAS mutations, Neoplasia
Hao, Kuz, Vorhies, Yan, Qiu, long-term modeling of SARS-cov-2 infection of in vitro cultured polarized human airway epithelium, mBio
Hornung, Köse-Vogel, Le Saux, Schulz, Radosa et al., Uncovering a unique pathogenic mechanism of SARS-cov-2 omicron variant: Selective induction of cellular senescence, Aging
Irizarry, Hobbs, Beazer-Barclay, Antonellis, Scherf, Speed, exploration, normalization, and summaries of high density oligonucleotide array probe level data, Biostatistics
Jamison, Zhou, Gelmann, Luk, Bates et al., entinostat in patients with relapsed or refractory abdominal neuroendocrine tumors, Oncologist
Jin, Du, Xu, Deng, Liu et al., Structure of M pro from SARS-cov-2 and discovery of its inhibitors, Nature
Jones, Sachs, Rouse, Wade, Shi, Multiple n-coR complexes contain distinct histone deacetylases, J. Biol. Chem
Knox, Wilson, Klinger, Franklin, Oler et al., DrugBank 6.0: the DrugBank knowledgebase for 2024, Nucleic Acids Res
Korotkevich, Sukhov, Budin, Shpak, Artyomov et al., None
Kretov, Clément, Lambert, Durand, N. Lyabin et al., YB-1, an abundant core mRnAbinding protein, has the capacity to form an RnA nucleoprotein filament: A structural analysis, Nucleic Acids Res
Liu, Lu, Plow, Qin, integrin mediates cell entry of the SARS-cov-2 virus independent of cellular receptor Ace2, J. Biol. Chem
Liu, Zou, Cui, Li, Wang et al., clinical hDAc inhibitors are effective drugs to prevent the entry of SARS-cov2, ACS Pharmacol. Transl. Sci
Maginnis, virus-receptor interactions: the key to cellular invasion, J. Mol. Biol
Malagola, Vasciaveo, Ochiai, Kim, Zheng et al., isthmus progenitor cells contribute to homeostatic cellular turnover and support regeneration following intestinal injury, Cell
Marcos-Villar, Pazo, Nieto, influenza virus and chromatin: Role of the chD1 chromatin remodeler in the virus life cycle, J. Virol
Margolin, Basso, Wiggins, Stolovitzky, Dalla Favera et al., ARAcne: An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context, BMC Bioinformatics
Martin, Amode, Aneja, Austine-Orimoloye, Azov et al., ensembl, Nucleic Acids Res
Melsted, Booeshaghi, Liu, Gao, Lu et al., Modular, efficient and constant-memory single-cell RnA-seq preprocessing, Nat. Biotechnol
Metz-Zumaran, Ramirez, Kee, Doldan, Shahraz et al., Single-cell analyses reveal SARS-cov-2 interference with intrinsic immune response in the human gut, Mol. Syst. Biol
Milacic, Beavers, Gong, Gillespie, Griss et al., D'eustachio, the reactome pathway knowledgebase 2024, Nucleic Acids Res
Mishra, Banerjea, SARS-cov-2 spike targets USP33-iRF9 axis via exosomal miR-148a to activate human microglia, Front. Immunol
Mishra, Re, Alvarez, Vasciaveo, Jacquier et al., Systematic elucidation of neuron-astrocyte interaction in models of amyotrophic lateral sclerosis using multi-modal integrated bioinformatics workflow, Nat. Commun
Mitrofanova, Bergren, Califano, Shen, A computational systems approach identifies synergistic specification genes that facilitate lineage conversion to prostate tissue, Nat. Commun
Mulay, Konda, Garcia, Yao, Beil et al., SARS-cov-2 infection of primary human lung epithelium for cOviD-19 modeling and drug discovery, Cell Rep
Mundi, Dela Cruz, Grunn, Diolaiti, Mauguen et al., A transcriptomebased precision oncology platform for patient-therapy alignment in a diverse set of treatment-resistant malignancies, Cancer Discov
Nakagawa, Tan, Alvarez, -E. Parfitt, Di Vicino et al., the Wnt-dependent master regulator nKX1-2 controls mouse pre-implantation development, Stem cell Reports
Obradovic, Ager, Khosravi-Maharlooei, Jackson, Yegnasubramanian et al., Systematic elucidation and pharmacological targeting of tumor-infiltrating regulatory t cell master regulators, Cancer Cell
Paull, Aytes, Jones, Subramaniam, Giorgi et al., A modular master regulator landscape controls cancer transcriptional identity, Cell
Polacco, Braberg, Fabius, Eckhardt, Soucheray et al., None
Program, Abdulla, Aevermann, Assis, Badajoz et al., cZ cellxGene Discover: A single-cell data platform for scalable exploration, analysis and modeling of aggregated data, Nucleic Acids Res
Puray-Chavez, N. Lee, Tenneti, Wang, Vuong, Liu et al., the translational landscape of SARS-cov-2-infected cells reveals suppression of innate immune genes, mBio
Qin, Xu, Wang, Jiang, Dai et al., Analyzing master regulators and scRnA-seq of cOviD-19 patients reveals an underlying anti-SARS-cov-2 mechanism of ZnF proteins, Brief. Bioinform
Rajbhandari, Salvatori, Yu, Rodriguez-Barrueco, Martinez et al., cross-cohort analysis identifies a teAD4-MYcn positive feedback loop as the core regulatory element of high-risk neuroblastoma, Cancer Discov
Ravindra, Alfajaro, Gasque, N. C. Huston, H. Wan, Szigeti-Buck et al., Single-cell longitudinal analysis of SARS-cov-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes, PLOS Biol
Rebendenne, Roy, Bonaventure, Desmarets, Arnaud-Arnould et al., Bidirectional genome-wide cRiSPR screens reveal host factors regulating SARS-cov-2, MeRS-cov and seasonal hcovs, Nat. Genet
Richter, Nayak, Taatjes, the mediator complex as a master regulator of transcription by RnA polymerase ii, Nat. Rev. Mol. Cell Biol
Rujas, Muthuraman, Liang, Landreth, Liao et al., Dual inhibition of vacuolar-AtPase and tMPRSS2 is required for complete blockade of SARS-cov-2 entry into cells, Antimicrob. Agents Chemother
Sayers, Cavanaugh, Clark, Pruitt, Schoch et al., None, GenBank. Nucleic Acids Res
Schneider, Luna, H.-H. Hoffmann, Sánchez-Rivera, Ashbrook, Le Pen et al., Genome-scale identification of SARS-cov-2 and pan-coronavirus host factor networks, Cell
Shaban, Müller, Mayr-Buro, Weiser, Schmitz et al., Reply to: the stress-inducible eR chaperone GRP78/BiP is upregulated during SARS-cov-2 infection and acts as a pro-viral protein, Nat. Commun
Silvestri, Vachani, Whitney, Elashoff, Smith et al., AeGiS Study team, A bronchial genomic classifier for the diagnostic evaluation of lung cancer, N. Engl. J. Med
Simons, Rinaldi, Bondu, Kell, Bradfute et al., integrin activation is an essential component of SARS-cov-2 infection, Sci. Rep
Son, Ding, Farb, Efanov, Sun et al., BAch2 inhibition reverses β cell failure in type 2 diabetes models, J. Clin. Invest
Stanifer, Bosker, Sun, Triana, Doldan et al., A model for network-based identification and pharmacological targeting of aberrant, replication-permissive transcriptional programs induced by viral infection, Commun. Biol
Study Team, Shared gene expression alterations in nasal and bronchial epithelium for lung cancer detection, J. Natl. Cancer Inst
Tang, Xue, Zhang, Nilsson-Payant, Duan et al., A multi-organoid platform identifies ciARt as a key factor for SARS-cov-2 infection, Nat. Cell Biol
Thankachan, Setty, KiF13A-A key regulator of recycling endosome dynamics, Front. Cell Dev. Biol
Tran, Grimley, Mcauley, Earnest, Wong et al., Air-liquid-interface differentiated human nose epithelium: A robust primary tissue culture model of SARS-cov-2 infection, Int. J. Mol. Sci
Underhill, Qutob, Yee, Torchia, A novel nuclear receptor corepressor complex, n-coR, contains components of the mammalian SWi/SnF complex and the corepressor KAP-1, J. Biol. Chem
Wang, -H. Lee, Xiong, Kim, Al Hasani et al., SARS-cov-2 restructures the host chromatin architecture, bioRxiv, doi:10.1101/2021.07.20.453146
Wei, Alfajaro, Deweirdt, Hanna, Lu-Culligan et al., Genome-wide cRiSPR screens reveal host factors critical for SARS-cov-2 infection, Cell
Wei, Patil, Collings, Alfajaro, Liang et al., Pharmacological disruption of mSWi/SnF complex activity restricts SARS-cov-2 infection, Nat. Genet
West, Johnstone, new and emerging hDAc inhibitors for cancer treatment, J. Clin. Invest
Wu, Lidsky, Xiao, Cheng, Jiang et al., SARS-cov-2 replication in airway epithelia requires motile cilia and microvillar reprogramming, Cell
Wyler, Mosbauer, Franke, Diag, Gottula et al., transcriptomic profiling of SARS-cov-2 infected human cell lines identifies hSP90 as target for cOviD-19 therapy, iScience
Xiang, Rir-Sim-Ah, tesfaigzi, il-9 and il-13 induce mucous cell metaplasia that is reduced by iFnγ in a Bax-mediated pathway, Am. J. Respir. Cell Mol. Biol
Yang, Jia, Meng, Xue, Liu et al., SnX27 suppresses SARS-cov-2 infection by inhibiting viral lysosome/late endosome entry, Proc. Natl. Acad. Sci. U.S.A
Yu, He, ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization, Mol. Biosyst
Zeleke, Pan, Onishi, Li, H. Tan, Alvarez et al., network-based assessment of hDAc6 activity predicts preclinical and clinical responses to the hDAc6 inhibitor ricolinostat in breast cancer, Nat. Cancer
Zhang, Yang, Roy, Liu, Roberts et al., tight junction protein occludin is an internalization factor for SARS-cov-2 infection and mediates virus cell-to-cell transmission, Proc. Natl. Acad. Sci. U.S.A
Zheng, Daskalogianni, Malbert-Colas, Karakostis, Sénéchal et al., the nascent polypeptide-associated complex (nAc) controls translation initiation in cis by recruiting nucleolin to the encoding mRnA, Nucleic Acids Res
Zhou, Chen, Liu, Liu, Liao et al., A cullin 5-based complex serves as an essential modulator of ORF9b stability in SARS-cov-2 replication, Signal Transduct. Target. Ther
Zhou, Liao, Liao, Liao, Lu, Ribosomal proteins: Functions beyond the ribosome, J. Mol. Cell Biol
Zhou, Yang, Guo, Qian, Ge et al., Airway basal cells show regionally distinct potential to undergo metaplastic differentiation, eLife
Zhu, Feng, Hu, Wang, Yu et al., A genome-wide cRiSPR screen identifies host factors that regulate SARS-cov-2 entry, Nat. Commun
DOI record:
{
"DOI": "10.1126/sciadv.adu2079",
"ISSN": [
"2375-2548"
],
"URL": "http://dx.doi.org/10.1126/sciadv.adu2079",
"abstract": "<jats:p>The impact of SARS-CoV-2 in the lung has been extensively studied, yet the molecular regulators of host-cell programs hijacked by the virus in distinct human airway epithelial cell populations remain poorly understood. Some of the reasons include overreliance on transcriptomic profiling and use of nonprimary cell systems. Here we report a network-based analysis of single-cell transcriptomic profiles able to identify master regulator (MR) proteins controlling SARS-CoV-2–mediated reprogramming in pathophysiologically relevant human ciliated, secretory, and basal cells. This underscored chromatin remodeling, endosomal sorting, ubiquitin pathways, as well as proviral factors identified by CRISPR assays as components of the viral-host response in these cells. Large-scale drug perturbation screens revealed 11 candidate drugs able to invert the entire MR signature activated by SARS-CoV-2. Leveraging MR analysis and perturbational profiles of human primary cells represents an innovative approach to investigate pathogen-host interactions in multiple airway conditions for drug prioritization.</jats:p>",
"alternative-id": [
"10.1126/sciadv.adu2079"
],
"assertion": [
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2024-10-29"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "2025-04-11"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 3,
"value": "2025-05-16"
}
],
"author": [
{
"ORCID": "https://orcid.org/0000-0002-3948-1548",
"affiliation": [
{
"name": "Columbia Center for Human Development, Columbia University Irving Medical Center, New York, NY 10032, USA."
},
{
"name": "Department of Genetics and Development, Columbia University Irving Medical Center, New York, NY 10032, USA."
}
],
"authenticated-orcid": true,
"family": "Dirvin",
"given": "Brooke",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0000-0002-9299-4852",
"affiliation": [
{
"name": "Department of Systems Biology, Columbia University Irving Medical Center, New York, NY 10032, USA."
},
{
"name": "Herbert Irving Comprehensive Cancer Center, Columbia University Irving Medical Center, New York, NY 10032, USA."
},
{
"name": "Institute for Systems Biology, Seattle, WA 98109, USA."
}
],
"authenticated-orcid": true,
"family": "Noh",
"given": "Heeju",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-7720-9166",
"affiliation": [
{
"name": "Department of Systems Biology, Columbia University Irving Medical Center, New York, NY 10032, USA."
},
{
"name": "Herbert Irving Comprehensive Cancer Center, Columbia University Irving Medical Center, New York, NY 10032, USA."
},
{
"name": "DarwinHealth Inc., New York, NY 10018, USA."
}
],
"authenticated-orcid": true,
"family": "Tomassoni",
"given": "Lorenzo",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0009-0003-7469-4033",
"affiliation": [
{
"name": "Columbia Center for Human Development, Columbia University Irving Medical Center, New York, NY 10032, USA."
},
{
"name": "Department of Medicine, Pulmonary Allergy Critical Care, Columbia University Irving Medical Center, New York, NY 10032, USA."
}
],
"authenticated-orcid": true,
"family": "Cao",
"given": "Danting",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-1903-0798",
"affiliation": [
{
"name": "Columbia Center for Human Development, Columbia University Irving Medical Center, New York, NY 10032, USA."
},
{
"name": "Department of Medicine, Pulmonary Allergy Critical Care, Columbia University Irving Medical Center, New York, NY 10032, USA."
}
],
"authenticated-orcid": true,
"family": "Zhou",
"given": "Yizhuo",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0009-0008-4923-1922",
"affiliation": [
{
"name": "Columbia Center for Human Development, Columbia University Irving Medical Center, New York, NY 10032, USA."
},
{
"name": "Department of Pharmacology, Columbia University Irving Medical Center, New York, NY 10032, USA."
}
],
"authenticated-orcid": true,
"family": "Ke",
"given": "Xiangyi",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0009-0009-4473-3912",
"affiliation": [
{
"name": "Columbia Center for Human Development, Columbia University Irving Medical Center, New York, NY 10032, USA."
},
{
"name": "Department of Medicine, Pulmonary Allergy Critical Care, Columbia University Irving Medical Center, New York, NY 10032, USA."
}
],
"authenticated-orcid": true,
"family": "Qian",
"given": "Jun",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-5671-1632",
"affiliation": [
{
"name": "Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA."
},
{
"name": "The Rockefeller University, New York, NY 10065, USA."
}
],
"authenticated-orcid": true,
"family": "Jangra",
"given": "Sonia",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-3156-3132",
"affiliation": [
{
"name": "Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA."
},
{
"name": "The Rockefeller University, New York, NY 10065, USA."
}
],
"authenticated-orcid": true,
"family": "Schotsaert",
"given": "Michael",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-6551-1827",
"affiliation": [
{
"name": "Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA."
}
],
"authenticated-orcid": true,
"family": "García-Sastre",
"given": "Adolfo",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-4012-8677",
"affiliation": [
{
"name": "Herbert Irving Comprehensive Cancer Center, Columbia University Irving Medical Center, New York, NY 10032, USA."
},
{
"name": "Department of Systems Biology, J.P. Sulzberger Columbia Genome Center, Columbia University Irving Medical Center, New York, NY 10032, USA."
}
],
"authenticated-orcid": true,
"family": "Karan",
"given": "Charles",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-4742-3679",
"affiliation": [
{
"name": "Herbert Irving Comprehensive Cancer Center, Columbia University Irving Medical Center, New York, NY 10032, USA."
},
{
"name": "DarwinHealth Inc., New York, NY 10018, USA."
},
{
"name": "Department of Systems Biology, J.P. Sulzberger Columbia Genome Center, Columbia University Irving Medical Center, New York, NY 10032, USA."
},
{
"name": "Department of Biomedical Informatics, Vagelos College of Physicians and Surgeons, Columbia University, New York, NY 10032, USA."
},
{
"name": "Department of Medicine, Vagelos College of Physicians and Surgeons, Columbia University, New York, NY 10032, USA."
},
{
"name": "Chan Zuckerberg Biohub New York, New York, NY, USA."
}
],
"authenticated-orcid": true,
"family": "Califano",
"given": "Andrea",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-8868-9716",
"affiliation": [
{
"name": "Columbia Center for Human Development, Columbia University Irving Medical Center, New York, NY 10032, USA."
},
{
"name": "Department of Genetics and Development, Columbia University Irving Medical Center, New York, NY 10032, USA."
},
{
"name": "Department of Medicine, Pulmonary Allergy Critical Care, Columbia University Irving Medical Center, New York, NY 10032, USA."
}
],
"authenticated-orcid": true,
"family": "Cardoso",
"given": "Wellington V.",
"sequence": "additional"
}
],
"container-title": "Science Advances",
"container-title-short": "Sci. Adv.",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"www.science.org"
]
},
"created": {
"date-parts": [
[
2025,
5,
16
]
],
"date-time": "2025-05-16T17:58:50Z",
"timestamp": 1747418330000
},
"deposited": {
"date-parts": [
[
2025,
5,
16
]
],
"date-time": "2025-05-16T17:59:22Z",
"timestamp": 1747418362000
},
"indexed": {
"date-parts": [
[
2025,
5,
17
]
],
"date-time": "2025-05-17T04:12:48Z",
"timestamp": 1747455168646,
"version": "3.40.5"
},
"is-referenced-by-count": 0,
"issue": "20",
"issued": {
"date-parts": [
[
2025,
5,
16
]
]
},
"journal-issue": {
"issue": "20",
"published-print": {
"date-parts": [
[
2025,
5,
16
]
]
}
},
"language": "en",
"link": [
{
"URL": "https://www.science.org/doi/pdf/10.1126/sciadv.adu2079",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "221",
"original-title": [],
"prefix": "10.1126",
"published": {
"date-parts": [
[
2025,
5,
16
]
]
},
"published-print": {
"date-parts": [
[
2025,
5,
16
]
]
},
"publisher": "American Association for the Advancement of Science (AAAS)",
"reference": [
{
"DOI": "10.1016/j.cell.2020.02.052",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_2_2"
},
{
"DOI": "10.1038/s41586-020-2286-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_3_2"
},
{
"DOI": "10.1038/s41588-022-01110-2",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_4_2"
},
{
"DOI": "10.1016/j.cell.2020.12.006",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_5_2"
},
{
"DOI": "10.15252/msb.202110232",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_6_2"
},
{
"DOI": "10.1016/j.cell.2020.10.028",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_7_2"
},
{
"DOI": "10.1016/j.isci.2021.102151",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_8_2"
},
{
"DOI": "10.1038/s41467-021-21213-4",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_9_2"
},
{
"DOI": "10.1038/s41556-023-01095-y",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_10_2"
},
{
"DOI": "10.1038/ng.3593",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_11_2"
},
{
"DOI": "10.1038/s41467-018-03843-3",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_12_2"
},
{
"article-title": "What is a master regulator?",
"author": "Chan S. S.-K.",
"first-page": "114",
"journal-title": "J. Stem Cell Res. Ther.",
"key": "e_1_3_2_13_2",
"unstructured": "S. S.-K. Chan, M. Kyba, What is a master regulator? J. Stem Cell Res. Ther. 3, 114 (2013).",
"volume": "3",
"year": "2013"
},
{
"DOI": "10.1016/j.cell.2020.11.045",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_14_2"
},
{
"DOI": "10.1186/1471-2105-7-S1-S7",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_15_2"
},
{
"DOI": "10.1038/nature08712",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_16_2"
},
{
"DOI": "10.1016/j.ccr.2014.03.017",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_17_2"
},
{
"DOI": "10.1158/2159-8290.CD-16-0861",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_18_2"
},
{
"DOI": "10.1158/2159-8290.CD-22-1020",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_19_2"
},
{
"DOI": "10.1038/s41588-018-0138-4",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_20_2"
},
{
"DOI": "10.1038/s43018-022-00489-5",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_21_2"
},
{
"DOI": "10.1093/oncolo/oyae118",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_22_2"
},
{
"DOI": "10.1016/j.celrep.2020.108474",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_23_2"
},
{
"DOI": "10.1172/JCI153876",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_24_2"
},
{
"DOI": "10.1038/ncomms14662",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_25_2"
},
{
"DOI": "10.1038/s41467-020-19177-y",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_26_2"
},
{
"DOI": "10.1016/j.celrep.2015.06.019",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_27_2"
},
{
"DOI": "10.1038/nn.4070",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_28_2"
},
{
"DOI": "10.1016/j.cell.2024.05.004",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_29_2"
},
{
"DOI": "10.1016/j.stemcr.2024.04.004",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_30_2"
},
{
"DOI": "10.1038/s42003-022-03663-8",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_31_2"
},
{
"DOI": "10.1038/s41588-022-01131-x",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_32_2"
},
{
"DOI": "10.1016/j.cell.2020.10.030",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_33_2"
},
{
"DOI": "10.1038/s41590-018-0276-y",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_34_2"
},
{
"DOI": "10.1371/journal.pbio.3001143",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_35_2"
},
{
"DOI": "10.1016/j.celrep.2021.109055",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_36_2"
},
{
"DOI": "10.1128/mbio.00815-22",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_37_2"
},
{
"DOI": "10.1056/NEJMoa1504601",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_38_2"
},
{
"article-title": "Shared gene expression alterations in nasal and bronchial epithelium for lung cancer detection",
"author": "AEGIS Study Team",
"first-page": "djw327",
"journal-title": "J. Natl. Cancer Inst.",
"key": "e_1_3_2_39_2",
"unstructured": "AEGIS Study Team, Shared gene expression alterations in nasal and bronchial epithelium for lung cancer detection. J. Natl. Cancer Inst. 109, djw327 (2017).",
"volume": "109",
"year": "2017"
},
{
"DOI": "10.1016/j.cels.2015.12.004",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_40_2"
},
{
"DOI": "10.3390/ijms23020835",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_41_2"
},
{
"DOI": "10.1093/bib/bbab118",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_42_2"
},
{
"DOI": "10.1128/mBio.02852-20",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_43_2"
},
{
"DOI": "10.1007/s00203-020-02094-5",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_44_2"
},
{
"DOI": "10.1093/jmcb/mjv014",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_45_2"
},
{
"DOI": "10.1093/nar/gkac751",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_46_2"
},
{
"DOI": "10.1093/nar/gky1303",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_47_2"
},
{
"DOI": "10.1158/0008-5472.CAN-17-0216",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_48_2"
},
{
"DOI": "10.1016/j.cell.2022.11.030",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_49_2"
},
{
"DOI": "10.1165/rcmb.2007-0078OC",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_50_2"
},
{
"DOI": "10.1073/pnas.2218623120",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_51_2"
},
{
"article-title": "How COVID-19 hijacks the cytoskeleton: Therapeutic implications",
"author": "Aminpour M.",
"first-page": "814",
"journal-title": "Life (Basel)",
"key": "e_1_3_2_52_2",
"unstructured": "M. Aminpour, S. Hameroff, J. A. Tuszynski, How COVID-19 hijacks the cytoskeleton: Therapeutic implications. Life (Basel) 12, 814 (2022).",
"volume": "12",
"year": "2022"
},
{
"DOI": "10.1016/j.jbc.2022.101710",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_53_2"
},
{
"DOI": "10.1016/j.jmb.2018.06.024",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_54_2"
},
{
"DOI": "10.1038/s41598-021-99893-7",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_55_2"
},
{
"DOI": "10.18632/aging.205297",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_56_2"
},
{
"DOI": "10.1038/s41580-022-00498-3",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_57_2"
},
{
"DOI": "10.1128/aac.00439-22",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_58_2"
},
{
"DOI": "10.1038/s41564-021-00958-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_59_2"
},
{
"DOI": "10.3389/fcell.2022.877532",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_60_2"
},
{
"DOI": "10.3389/fimmu.2021.656700",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_61_2"
},
{
"DOI": "10.1038/s41392-024-01874-5",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_62_2"
},
{
"DOI": "10.1073/pnas.2117576119",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_63_2"
},
{
"DOI": "10.1038/s41588-023-01307-z",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_64_2"
},
{
"DOI": "10.1158/2159-8290.CD-22-0342",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_65_2"
},
{
"DOI": "10.1016/j.ccell.2023.04.003",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_66_2"
},
{
"DOI": "10.1093/nar/gkad976",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_67_2"
},
{
"DOI": "10.1021/acsptsci.0c00163",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_68_2"
},
{
"DOI": "10.1128/JVI.00053-16",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_69_2"
},
{
"DOI": "10.1016/j.neo.2022.100837",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_70_2"
},
{
"DOI": "10.1038/s41467-022-34066-2",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_71_2"
},
{
"DOI": "10.1038/s41467-017-00136-z",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_72_2"
},
{
"DOI": "10.1056/NEJMoa0808427",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_73_2"
},
{
"DOI": "10.1093/nar/gkad1025",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_74_2"
},
{
"DOI": "10.1074/jbc.C000879200",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_75_2"
},
{
"DOI": "10.1074/jbc.M007864200",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_76_2"
},
{
"DOI": "10.1101/2021.07.20.453146",
"doi-asserted-by": "crossref",
"key": "e_1_3_2_77_2",
"unstructured": "R. Wang J.-H. Lee F. Xiong J. Kim L. Al Hasani X. Yuan P. Shivshankar J. Krakowiak C. Qi Y. Wang H. K. Eltzschig W. Li SARS-CoV-2 restructures the host chromatin architecture. bioRxiv 453146 [Preprint] (2021). https://doi.org/10.1101/2021.07.20.453146."
},
{
"DOI": "10.1172/JCI69738",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_78_2"
},
{
"DOI": "10.1038/s41586-020-2223-y",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_79_2"
},
{
"DOI": "10.1016/j.isci.2023.107374",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_80_2"
},
{
"DOI": "10.7554/eLife.80083",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_81_2"
},
{
"DOI": "10.1093/nar/gkaa1023",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_82_2"
},
{
"DOI": "10.1038/s41587-021-00870-2",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_83_2"
},
{
"DOI": "10.1093/bioinformatics/btw216",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_84_2"
},
{
"DOI": "10.1093/biostatistics/4.2.249",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_85_2"
},
{
"DOI": "10.1093/nar/gkae1142",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_86_2"
},
{
"key": "e_1_3_2_87_2",
"unstructured": "G. Korotkevich V. Sukhov N. Budin B. Shpak M. Artyomov A. Sergushichev. (bioRxiv 2016)."
},
{
"DOI": "10.1038/nbt.3519",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_88_2"
},
{
"DOI": "10.1093/nar/gkac958",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_89_2"
},
{
"DOI": "10.1039/C5MB00663E",
"doi-asserted-by": "publisher",
"key": "e_1_3_2_90_2"
}
],
"reference-count": 89,
"references-count": 89,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.science.org/doi/10.1126/sciadv.adu2079"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Identification and targeting of regulators of SARS-CoV-2–host interactions in the airway epithelium",
"type": "journal-article",
"update-policy": "https://doi.org/10.34133/aaas_crossmark",
"volume": "11"
}