Stem loop binding protein promotes SARS-CoV-2 replication via -1 programmed ribosomal frameshifting
et al., Signal Transduction and Targeted Therapy, doi:10.1038/s41392-025-02277-w, Jun 2025
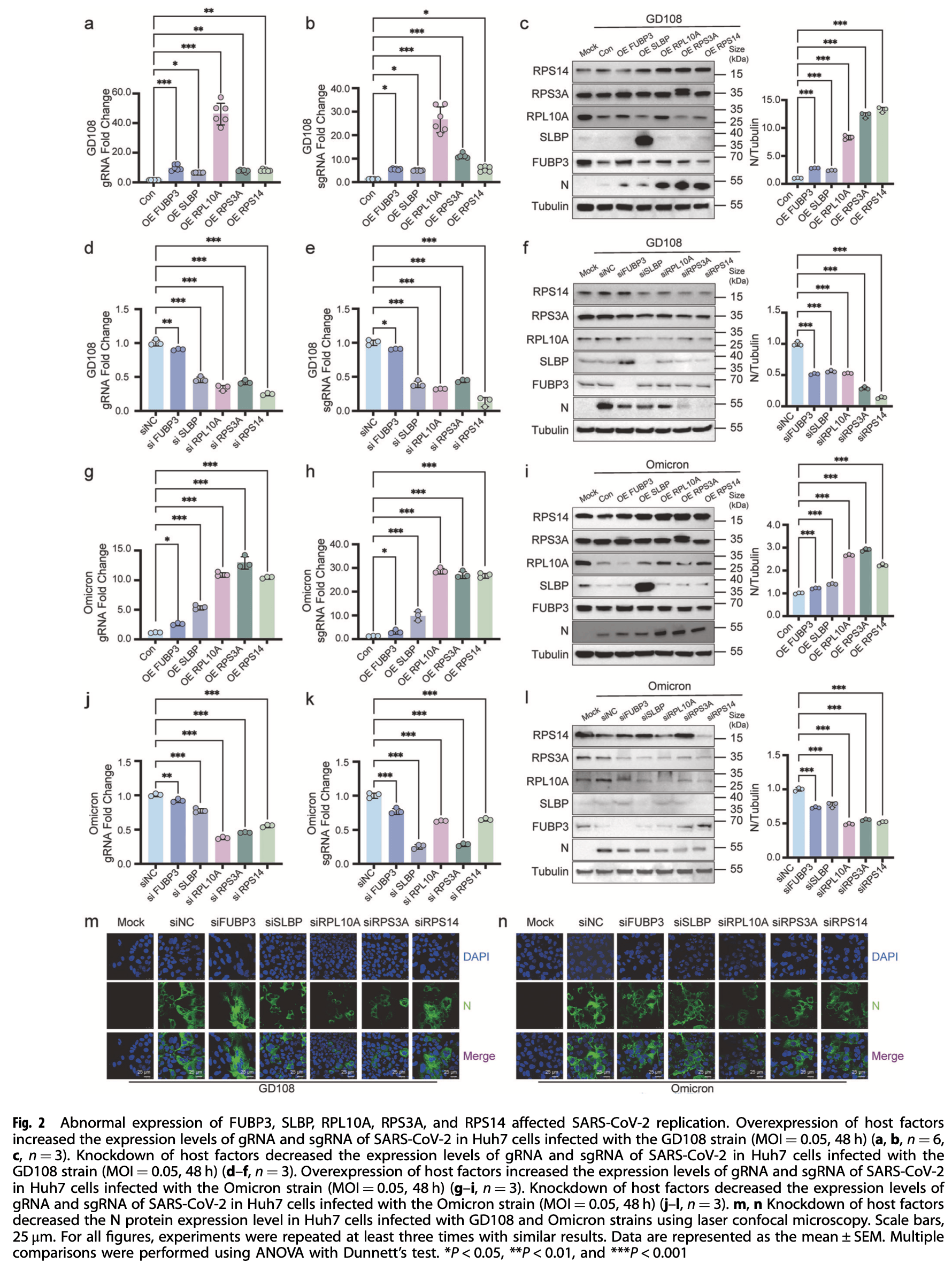
In vitro study identifying SLBP (Stem Loop Binding Protein) as a host factor that promotes SARS-CoV-2 replication by enhancing -1 programmed ribosomal frameshifting (-1 PRF). Authors used RNA pull-down assays with mass spectrometry to identify five host proteins (FUBP3, SLBP, RPL10A, RPS3A, and RPS14) that interact with -1 PRF RNA. SLBP overexpression enhanced viral replication while its knockdown reduced viral propagation in Huh7 and H1299 cells.
Chen et al., 13 Jun 2025, peer-reviewed, 12 authors.
Contact: yunpeng1987@hotmail.com, lushuaiyao-km@163.com, pengxiaozhong@pumc.edu.cn.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Stem loop binding protein promotes SARS-CoV-2 replication via -1 programmed ribosomal frameshifting
Signal Transduction and Targeted Therapy, doi:10.1038/s41392-025-02277-w
The -1 programmed ribosomal frameshifting (-1 PRF) in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is crucial for keeping the balance between pp1a and pp1ab polyproteins. To date, the host factors influencing this process remain poorly understood. Using RNA pull-down assays combined with mass spectrometry screening, we discovered five host proteins interacting with -1 PRF RNA, including Stem Loop Binding Protein (SLBP). Our findings revealed that SLBP overexpression enhanced frameshifting and promoted viral replication. Moreover, the interaction between SLBP and -1 PRF RNA was predicted using the PrismNet deep learning tool, which calculated a high binding probability of 0.922. Using Electrophoretic Mobility Shift Assays (EMSAs) and RNA pull down assays, our findings demonstrated SLBP's direct binding to the SARS-CoV-2 genome, with preferential affinity for the stem loop 3 region of the -1 PRF RNA. Using smFISH assays, we further confirmed their physical colocalization. The role of SLBP in promoting frameshifting was verified using an in vitro translation system. Further investigation showed that SLBP deletions reshaped the host factor pattern around -1 PRF RNA, diminishing interactions with FUBP3 and RPS3A while enhancing RPL10A binding. Together, our findings identify SLBP as a host protein that promotes SARS-CoV-2 frameshifting, highlighting its potential as a druggable target for COVID-19.
AUTHOR CONTRIBUTIONS X.Z.P. and T.X.C. designed the experiments. T.X.C. performed the most experiments, analyzed the data, and wrote the manuscript. R.M.Z. and T.F.D. performed virus infection experiments. Y.P.L. performed the EMSA and Native-PAGE assays. H.Y. and X.T.Z. helped with methodology, and software. Z.X.W. helped with bioinformatics analysis. Y.Z., W.Q.Q., and B.Y. provided reagents and cell lines. X.Z.P., S.Y.L., and Y.P.L. provided comments, and supervised the project. All authors have read and approved the article.
ADDITIONAL INFORMATION Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41392-025-02277-w . Competing interests: The authors declare no competing interests. Publisher's note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Albright, Human cytomegalovirus lytic infection inhibits replicationdependent histone synthesis and requires stem loop binding protein function, Proc. Natl. Acad. Sci. USA
Belew, Ribosomal frameshifting in the CCR5 mRNA is regulated by miRNAs and the NMD pathway, Nature
Bhatt, Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome, Science
Brierley, Digard, Inglis, Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot, Cell
Carmody, Coordination of -1 programmed ribosomal frameshifting by transcript and nascent chain features revealed by deep mutational scanning, Nucleic Acids Res
Ceylan, Optimization of structure-guided development of chemical probes for the Pseudoknot RNA of the frameshift element in SARS-CoV-2, Angew. Chem. Int. Ed. Engl
Chang, Transcriptional and epi-transcriptional dynamics of SARS-CoV-2 during cellular infection, Cell Rep
Dong, FUBP3 degrades the Porcine epidemic diarrhea virus nucleocapsid protein and induces the production of type I interferon, J. Virol
Fan, RNA binding motif 4 inhibits the replication of ebolavirus by directly targeting 3'-leader region of genomic RNA, Emerg. Microbes Infect
Grifoni, SARS-CoV-2 human T cell epitopes: adaptive immune response against COVID-19, Cell Host Microbe
Iioka, Loiselle, Haystead, Macara, Efficient detection of RNAprotein interactions using tethered RNAs, Nucleic Acids Res
Jacks, Varmus, Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting, Science
Katahira, Nsp14 of SARS-CoV-2 inhibits mRNA processing and nuclear export by targeting the nuclear cap-binding complex, Nucleic Acids Res
Kelly, Structural and functional conservation of the programmed -1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2), J. Biol. Chem
Khatter, Myasnikov, Natchiar, Klaholz, Structure of the human 80S ribosome, Nature
Lee, Absolute quantitation of individual SARS-CoV-2 RNA molecules provides a new paradigm for infection dynamics and variant sdifferences, Elife
Li, Carrimycin inhibits coronavirus replication by decreasing the efficiency of programmed -1 ribosomal frameshifting through directly binding to the RNA pseudoknot of viral frameshift-stimulatory element, Acta Pharm. Sin. B
Li, Stem-loop binding protein is a multifaceted cellular regulator of HIV-1 replication, J. Clin. Investig
Manktelow, Shigemoto, Brierley, Characterization of the frameshift signal of Edr, a mammalian example of programmed -1 ribosomal frameshifting, Nucleic Acids Res
Mei, Kosakovsky, Pond, Nekrutenko, Stepwise Evolution and exceptional conservation of ORF1a/b overlap in coronaviruses, Mol. Biol. Evol
Napthine, A novel role for poly(C) binding proteins in programmed ribosomal frameshifting, Nucleic Acids Res
Napthine, Hill, Nugent, Brierley, Modulation of viral programmed ribosomal frameshifting and stop codon readthrough by the host restriction factor shiftless, Viruses
Patel, Molecular characterization of the RNA-protein complex directing -2/-1 programmed ribosomal frameshifting during arterivirus replicase expression, J. Biol. Chem
Paternina-Caicedo, Effectiveness of CoronaVac and BNT162b2 COVID-19 mass vaccination in Colombia: a population-based cohort study, Lancet Reg. Health Am
Rodriguez, Srivastav, Muller, C19ORF66 broadly escapes virus-induced endonuclease cleavage and restricts Kaposi's Sarcoma-Associated Herpesvirus, J. Virol
Schmidt, The SARS-CoV-2 RNA-protein interactome in infected human cells, Nat. Microbiol
Sun, Restriction of SARS-CoV-2 replication by targeting programmed -1 ribosomal frameshifting, Proc. Natl. Acad. Sci. USA
Suzuki, Characterization of RyDEN (C19orf66) as an interferon-stimulated cellular inhibitor against dengue virus replication, PLoS Pathog
Tan, Marzluff, Dominski, Tong, Structure of histone mRNA stemloop, human stem-loop binding protein, and 3'hExo ternary complex, Science
Tsai, Kaempferol inhibits enterovirus 71 replication and internal ribosome entry site (IRES) activity through FUBP and HNRP proteins, Food Chem
Varricchio, Geneticin shows selective antiviral activity against SARS-CoV-2 by interfering with programmed -1 ribosomal frameshifting, Antivir. Res
Wang, Chen, Xiao, Zhao, Feng, Rapid COVID-19 rebound in a severe COVID-19 patient during 20-day course of Paxlovid, J. Infect
Wang, Regulation of HIV-1 Gag-Pol expression by shiftless, an inhibitor of programmed -1 ribosomal frameshifting, Cell
Wei, Sun, Guo, Genome-wide CRISPR screens identify noncanonical translation factor eIF2A as an enhancer of SARS-CoV-2 programmed -1 ribosomal frameshifting, Cell Rep
Williams, Marzluff, The sequence of the stem and flanking sequences at the 3' end of histone mRNA are critical determinants for the binding of the stem-loop binding protein, Nucleic Acids Res
Wills, Moore, Hammer, Gesteland, Atkins, A functional -1 ribosomal frameshift signal in the human paraneoplastic Ma3 gene, J. Biol. Chem
Xu, PrismNet: predicting protein-RNA interaction using in vivo RNA structural information, Nucleic Acids Res
Xu, Tong, Chen, FUSE binding protein FUBP3 is a potent regulator in Japanese encephalitis virus infection, Virol. J
Yu, C19orf66 inhibits japanese encephalitis virus replication by targeting -1 PRF and the NS3 protein, Virol. Sin
Yu, Pseudoknot-targeting Cas13b combats SARS-CoV-2 infection by suppressing viral replication, Mol. Ther
Zhang, Capturing RNA-protein interaction via CRUIS, Nucleic Acids Res
Zhou, Therapeutic targets and interventional strategies in COVID-19: mechanisms and clinical studies, Signal. Transduct. Target Ther
Zimmer, The short isoform of the host antiviral protein ZAP acts as an inhibitor of SARS-CoV-2 programmed ribosomal frameshifting, Nat. Commun
DOI record:
{
"DOI": "10.1038/s41392-025-02277-w",
"ISSN": [
"2059-3635"
],
"URL": "http://dx.doi.org/10.1038/s41392-025-02277-w",
"abstract": "<jats:title>Abstract</jats:title>\n <jats:p>The -1 programmed ribosomal frameshifting (-1 PRF) in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is crucial for keeping the balance between pp1a and pp1ab polyproteins. To date, the host factors influencing this process remain poorly understood. Using RNA pull-down assays combined with mass spectrometry screening, we discovered five host proteins interacting with -1 PRF RNA, including Stem Loop Binding Protein (SLBP). Our findings revealed that SLBP overexpression enhanced frameshifting and promoted viral replication. Moreover, the interaction between SLBP and -1 PRF RNA was predicted using the PrismNet deep learning tool, which calculated a high binding probability of 0.922. Using Electrophoretic Mobility Shift Assays (EMSAs) and RNA pull down assays, our findings demonstrated SLBP’s direct binding to the SARS-CoV-2 genome, with preferential affinity for the stem loop 3 region of the -1 PRF RNA. Using smFISH assays, we further confirmed their physical colocalization. The role of SLBP in promoting frameshifting was verified using an in vitro translation system. Further investigation showed that SLBP deletions reshaped the host factor pattern around -1 PRF RNA, diminishing interactions with FUBP3 and RPS3A while enhancing RPL10A binding. Together, our findings identify SLBP as a host protein that promotes SARS-CoV-2 frameshifting, highlighting its potential as a druggable target for COVID-19.</jats:p>",
"alternative-id": [
"2277"
],
"article-number": "192",
"assertion": [
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Received",
"name": "received",
"order": 1,
"value": "6 July 2023"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Revised",
"name": "revised",
"order": 2,
"value": "24 April 2025"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Accepted",
"name": "accepted",
"order": 3,
"value": "19 May 2025"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "First Online",
"name": "first_online",
"order": 4,
"value": "13 June 2025"
},
{
"group": {
"label": "Competing interests",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 1,
"value": "The authors declare no competing interests."
}
],
"author": [
{
"ORCID": "https://orcid.org/0000-0003-4877-9423",
"affiliation": [],
"authenticated-orcid": false,
"family": "Chen",
"given": "Tanxiu",
"sequence": "first"
},
{
"affiliation": [],
"family": "Zhu",
"given": "Ruimin",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-9814-0081",
"affiliation": [],
"authenticated-orcid": false,
"family": "Du",
"given": "Tingfu",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Yang",
"given": "Hao",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zhang",
"given": "Xintian",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wang",
"given": "Zhixing",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zhang",
"given": "Yong",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Quan",
"given": "Wenqi",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Yin",
"given": "Bin",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Liu",
"given": "Yunpeng",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-1675-9735",
"affiliation": [],
"authenticated-orcid": false,
"family": "Lu",
"given": "Shuaiyao",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-8564-0247",
"affiliation": [],
"authenticated-orcid": false,
"family": "Peng",
"given": "Xiaozhong",
"sequence": "additional"
}
],
"container-title": "Signal Transduction and Targeted Therapy",
"container-title-short": "Sig Transduct Target Ther",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"link.springer.com"
]
},
"created": {
"date-parts": [
[
2025,
6,
13
]
],
"date-time": "2025-06-13T12:35:50Z",
"timestamp": 1749818150000
},
"deposited": {
"date-parts": [
[
2025,
6,
13
]
],
"date-time": "2025-06-13T14:02:11Z",
"timestamp": 1749823331000
},
"indexed": {
"date-parts": [
[
2025,
6,
14
]
],
"date-time": "2025-06-14T04:06:17Z",
"timestamp": 1749873977670,
"version": "3.41.0"
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2025,
6,
13
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2025,
12
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
6,
13
]
],
"date-time": "2025-06-13T00:00:00Z",
"timestamp": 1749772800000
}
},
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
6,
13
]
],
"date-time": "2025-06-13T00:00:00Z",
"timestamp": 1749772800000
}
}
],
"link": [
{
"URL": "https://www.nature.com/articles/s41392-025-02277-w.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41392-025-02277-w",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41392-025-02277-w.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"prefix": "10.1038",
"published": {
"date-parts": [
[
2025,
6,
13
]
]
},
"published-online": {
"date-parts": [
[
2025,
6,
13
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference": [
{
"DOI": "10.1038/s41392-021-00733-x",
"author": "YW Zhou",
"doi-asserted-by": "publisher",
"first-page": "317",
"journal-title": "Signal. Transduct. Target Ther.",
"key": "2277_CR1",
"unstructured": "Zhou, Y. W. et al. Therapeutic targets and interventional strategies in COVID-19: mechanisms and clinical studies. Signal. Transduct. Target Ther. 6, 317 (2021).",
"volume": "6",
"year": "2021"
},
{
"DOI": "10.1016/j.jinf.2022.08.012",
"author": "Y Wang",
"doi-asserted-by": "publisher",
"first-page": "e134",
"journal-title": "J. Infect.",
"key": "2277_CR2",
"unstructured": "Wang, Y., Chen, X., Xiao, W., Zhao, D. & Feng, L. Rapid COVID-19 rebound in a severe COVID-19 patient during 20-day course of Paxlovid. J. Infect. 85, e134–e136 (2022).",
"volume": "85",
"year": "2022"
},
{
"DOI": "10.1093/nar/gkab1172",
"author": "PJ Carmody",
"doi-asserted-by": "publisher",
"first-page": "12943",
"journal-title": "Nucleic Acids Res.",
"key": "2277_CR3",
"unstructured": "Carmody, P. J. et al. Coordination of -1 programmed ribosomal frameshifting by transcript and nascent chain features revealed by deep mutational scanning. Nucleic Acids Res. 49, 12943–12954 (2021).",
"volume": "49",
"year": "2021"
},
{
"DOI": "10.1093/nar/gki299",
"author": "E Manktelow",
"doi-asserted-by": "publisher",
"first-page": "1553",
"journal-title": "Nucleic Acids Res.",
"key": "2277_CR4",
"unstructured": "Manktelow, E., Shigemoto, K. & Brierley, I. Characterization of the frameshift signal of Edr, a mammalian example of programmed -1 ribosomal frameshifting. Nucleic Acids Res. 33, 1553–1563 (2005).",
"volume": "33",
"year": "2005"
},
{
"DOI": "10.1074/jbc.M511629200",
"author": "NM Wills",
"doi-asserted-by": "publisher",
"first-page": "7082",
"journal-title": "J. Biol. Chem.",
"key": "2277_CR5",
"unstructured": "Wills, N. M., Moore, B., Hammer, A., Gesteland, R. F. & Atkins, J. F. A functional -1 ribosomal frameshift signal in the human paraneoplastic Ma3 gene. J. Biol. Chem. 281, 7082–7088 (2006).",
"volume": "281",
"year": "2006"
},
{
"DOI": "10.1038/nature13429",
"author": "AT Belew",
"doi-asserted-by": "publisher",
"first-page": "265",
"journal-title": "Nature",
"key": "2277_CR6",
"unstructured": "Belew, A. T. et al. Ribosomal frameshifting in the CCR5 mRNA is regulated by miRNAs and the NMD pathway. Nature 512, 265–269 (2014).",
"volume": "512",
"year": "2014"
},
{
"DOI": "10.1126/science.2416054",
"author": "T Jacks",
"doi-asserted-by": "publisher",
"first-page": "1237",
"journal-title": "Science",
"key": "2277_CR7",
"unstructured": "Jacks, T. & Varmus, H. E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science 230, 1237–1242 (1985).",
"volume": "230",
"year": "1985"
},
{
"DOI": "10.1016/0092-8674(89)90124-4",
"author": "I Brierley",
"doi-asserted-by": "publisher",
"first-page": "537",
"journal-title": "Cell",
"key": "2277_CR8",
"unstructured": "Brierley, I., Digard, P. & Inglis, S. C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell 57, 537–547 (1989).",
"volume": "57",
"year": "1989"
},
{
"DOI": "10.1093/molbev/msab265",
"author": "H Mei",
"doi-asserted-by": "publisher",
"first-page": "5678",
"journal-title": "Mol. Biol. Evol.",
"key": "2277_CR9",
"unstructured": "Mei, H., Kosakovsky Pond, S. & Nekrutenko, A. Stepwise Evolution and exceptional conservation of ORF1a/b overlap in coronaviruses. Mol. Biol. Evol. 38, 5678–5684 (2021).",
"volume": "38",
"year": "2021"
},
{
"DOI": "10.1073/pnas.2023051118",
"author": "Y Sun",
"doi-asserted-by": "publisher",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "2277_CR10",
"unstructured": "Sun, Y. et al. Restriction of SARS-CoV-2 replication by targeting programmed -1 ribosomal frameshifting. Proc. Natl. Acad. Sci. USA 118, e2023051118 (2021).",
"volume": "118",
"year": "2021"
},
{
"DOI": "10.1016/j.apsb.2024.02.023",
"author": "H Li",
"doi-asserted-by": "publisher",
"first-page": "2567",
"journal-title": "Acta Pharm. Sin. B",
"key": "2277_CR11",
"unstructured": "Li, H. et al. Carrimycin inhibits coronavirus replication by decreasing the efficiency of programmed -1 ribosomal frameshifting through directly binding to the RNA pseudoknot of viral frameshift-stimulatory element. Acta Pharm. Sin. B 14, 2567–2580 (2024).",
"volume": "14",
"year": "2024"
},
{
"DOI": "10.1016/j.celrep.2021.109108",
"author": "JJ Chang",
"doi-asserted-by": "publisher",
"first-page": "109108",
"journal-title": "Cell Rep.",
"key": "2277_CR12",
"unstructured": "Chang, J. J. et al. Transcriptional and epi-transcriptional dynamics of SARS-CoV-2 during cellular infection. Cell Rep. 35, 109108 (2021).",
"volume": "35",
"year": "2021"
},
{
"DOI": "10.1016/j.ymthe.2023.03.018",
"author": "D Yu",
"doi-asserted-by": "publisher",
"first-page": "1675",
"journal-title": "Mol. Ther.",
"key": "2277_CR13",
"unstructured": "Yu, D. et al. Pseudoknot-targeting Cas13b combats SARS-CoV-2 infection by suppressing viral replication. Mol. Ther. 31, 1675–1687 (2023).",
"volume": "31",
"year": "2023"
},
{
"DOI": "10.1126/science.abf3546",
"author": "PR Bhatt",
"doi-asserted-by": "publisher",
"first-page": "1306",
"journal-title": "Science",
"key": "2277_CR14",
"unstructured": "Bhatt, P. R. et al. Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. Science 372, 1306–1313 (2021).",
"volume": "372",
"year": "2021"
},
{
"DOI": "10.1002/anie.202417961",
"author": "B Ceylan",
"doi-asserted-by": "publisher",
"first-page": "e202417961",
"journal-title": "Angew. Chem. Int. Ed. Engl.",
"key": "2277_CR15",
"unstructured": "Ceylan, B. et al. Optimization of structure-guided development of chemical probes for the Pseudoknot RNA of the frameshift element in SARS-CoV-2. Angew. Chem. Int. Ed. Engl. 64, e202417961 (2025).",
"volume": "64",
"year": "2025"
},
{
"DOI": "10.1074/jbc.AC120.013449",
"author": "JA Kelly",
"doi-asserted-by": "publisher",
"first-page": "10741",
"journal-title": "J. Biol. Chem.",
"key": "2277_CR16",
"unstructured": "Kelly, J. A. et al. Structural and functional conservation of the programmed -1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2). J. Biol. Chem. 295, 10741–10748 (2020).",
"volume": "295",
"year": "2020"
},
{
"DOI": "10.1093/nar/gkaa143",
"author": "Z Zhang",
"doi-asserted-by": "publisher",
"journal-title": "Nucleic Acids Res.",
"key": "2277_CR17",
"unstructured": "Zhang, Z. et al. Capturing RNA-protein interaction via CRUIS. Nucleic Acids Res. 48, e52 (2020).",
"volume": "48",
"year": "2020"
},
{
"DOI": "10.3390/v13071230",
"author": "S Napthine",
"doi-asserted-by": "publisher",
"first-page": "1230",
"journal-title": "Viruses",
"key": "2277_CR18",
"unstructured": "Napthine, S., Hill, C. H., Nugent, H. C. M. & Brierley, I. Modulation of viral programmed ribosomal frameshifting and stop codon readthrough by the host restriction factor shiftless. Viruses 13, 1230 (2021).",
"volume": "13",
"year": "2021"
},
{
"DOI": "10.1038/s41467-021-27431-0",
"author": "MM Zimmer",
"doi-asserted-by": "publisher",
"journal-title": "Nat. Commun.",
"key": "2277_CR19",
"unstructured": "Zimmer, M. M. et al. The short isoform of the host antiviral protein ZAP acts as an inhibitor of SARS-CoV-2 programmed ribosomal frameshifting. Nat. Commun. 12, 7193 (2021).",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1016/j.celrep.2023.112987",
"author": "LH Wei",
"doi-asserted-by": "publisher",
"first-page": "112987",
"journal-title": "Cell Rep.",
"key": "2277_CR20",
"unstructured": "Wei, L. H., Sun, Y. & Guo, J. U. Genome-wide CRISPR screens identify noncanonical translation factor eIF2A as an enhancer of SARS-CoV-2 programmed -1 ribosomal frameshifting. Cell Rep. 42, 112987 (2023).",
"volume": "42",
"year": "2023"
},
{
"DOI": "10.1093/nar/gkq1316",
"author": "H Iioka",
"doi-asserted-by": "publisher",
"first-page": "e53",
"journal-title": "Nucleic Acids Res.",
"key": "2277_CR21",
"unstructured": "Iioka, H., Loiselle, D., Haystead, T. A. & Macara, I. G. Efficient detection of RNA-protein interactions using tethered RNAs. Nucleic Acids Res. 39, e53 (2011).",
"volume": "39",
"year": "2011"
},
{
"DOI": "10.1038/nature14427",
"author": "H Khatter",
"doi-asserted-by": "publisher",
"first-page": "640",
"journal-title": "Nature",
"key": "2277_CR22",
"unstructured": "Khatter, H., Myasnikov, A. G., Natchiar, S. K. & Klaholz, B. P. Structure of the human 80S ribosome. Nature 520, 640–645 (2015).",
"volume": "520",
"year": "2015"
},
{
"DOI": "10.1016/j.foodchem.2011.03.022",
"author": "FJ Tsai",
"doi-asserted-by": "publisher",
"first-page": "312",
"journal-title": "Food Chem.",
"key": "2277_CR23",
"unstructured": "Tsai, F. J. et al. Kaempferol inhibits enterovirus 71 replication and internal ribosome entry site (IRES) activity through FUBP and HNRP proteins. Food Chem. 128, 312–322 (2011).",
"volume": "128",
"year": "2011"
},
{
"DOI": "10.1128/jvi.00618-22",
"author": "S Dong",
"doi-asserted-by": "publisher",
"journal-title": "J. Virol.",
"key": "2277_CR24",
"unstructured": "Dong, S. et al. FUBP3 degrades the Porcine epidemic diarrhea virus nucleocapsid protein and induces the production of type I interferon. J. Virol. 96, e0061822 (2022).",
"volume": "96",
"year": "2022"
},
{
"DOI": "10.1073/pnas.2122174119",
"author": "ER Albright",
"doi-asserted-by": "publisher",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "2277_CR25",
"unstructured": "Albright, E. R. et al. Human cytomegalovirus lytic infection inhibits replication-dependent histone synthesis and requires stem loop binding protein function. Proc. Natl. Acad. Sci. USA 119, e2122174119 (2022).",
"volume": "119",
"year": "2022"
},
{
"DOI": "10.1186/s12985-021-01697-8",
"author": "P Xu",
"doi-asserted-by": "publisher",
"first-page": "224",
"journal-title": "Virol. J.",
"key": "2277_CR26",
"unstructured": "Xu, P., Tong, W. & Chen, Y. M. FUSE binding protein FUBP3 is a potent regulator in Japanese encephalitis virus infection. Virol. J. 18, 224 (2021).",
"volume": "18",
"year": "2021"
},
{
"DOI": "10.1172/JCI82360",
"author": "M Li",
"doi-asserted-by": "publisher",
"first-page": "3117",
"journal-title": "J. Clin. Investig.",
"key": "2277_CR27",
"unstructured": "Li, M. et al. Stem-loop binding protein is a multifaceted cellular regulator of HIV-1 replication. J. Clin. Investig. 126, 3117–3129 (2016).",
"volume": "126",
"year": "2016"
},
{
"DOI": "10.1093/nar/gkad483",
"author": "J Katahira",
"doi-asserted-by": "publisher",
"first-page": "7602",
"journal-title": "Nucleic Acids Res.",
"key": "2277_CR28",
"unstructured": "Katahira, J. et al. Nsp14 of SARS-CoV-2 inhibits mRNA processing and nuclear export by targeting the nuclear cap-binding complex. Nucleic Acids Res. 51, 7602–7618 (2023).",
"volume": "51",
"year": "2023"
},
{
"DOI": "10.1080/22221751.2023.2300762",
"author": "L Fan",
"doi-asserted-by": "publisher",
"journal-title": "Emerg. Microbes Infect.",
"key": "2277_CR29",
"unstructured": "Fan, L. et al. RNA binding motif 4 inhibits the replication of ebolavirus by directly targeting 3’-leader region of genomic RNA. Emerg. Microbes Infect. 13, 2300762 (2024).",
"volume": "13",
"year": "2024"
},
{
"DOI": "10.1016/j.antiviral.2022.105452",
"author": "C Varricchio",
"doi-asserted-by": "publisher",
"first-page": "105452",
"journal-title": "Antivir. Res.",
"key": "2277_CR30",
"unstructured": "Varricchio, C. et al. Geneticin shows selective antiviral activity against SARS-CoV-2 by interfering with programmed -1 ribosomal frameshifting. Antivir. Res. 208, 105452 (2022).",
"volume": "208",
"year": "2022"
},
{
"DOI": "10.1093/nar/23.4.654",
"author": "AS Williams",
"doi-asserted-by": "publisher",
"first-page": "654",
"journal-title": "Nucleic Acids Res.",
"key": "2277_CR31",
"unstructured": "Williams, A. S. & Marzluff, W. F. The sequence of the stem and flanking sequences at the 3’ end of histone mRNA are critical determinants for the binding of the stem-loop binding protein. Nucleic Acids Res. 23, 654–662 (1995).",
"volume": "23",
"year": "1995"
},
{
"DOI": "10.1126/science.1228705",
"author": "D Tan",
"doi-asserted-by": "publisher",
"first-page": "318",
"journal-title": "Science",
"key": "2277_CR32",
"unstructured": "Tan, D., Marzluff, W. F., Dominski, Z. & Tong, L. Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3’hExo ternary complex. Science 339, 318–321 (2013).",
"volume": "339",
"year": "2013"
},
{
"DOI": "10.1016/j.chom.2022.10.017",
"author": "A Grifoni",
"doi-asserted-by": "publisher",
"first-page": "1788",
"journal-title": "Cell Host Microbe",
"key": "2277_CR33",
"unstructured": "Grifoni, A. et al. SARS-CoV-2 human T cell epitopes: adaptive immune response against COVID-19. Cell Host Microbe 30, 1788 (2022).",
"volume": "30",
"year": "2022"
},
{
"author": "A Paternina-Caicedo",
"journal-title": "Lancet Reg. Health Am.",
"key": "2277_CR34",
"unstructured": "Paternina-Caicedo, A. et al. Effectiveness of CoronaVac and BNT162b2 COVID-19 mass vaccination in Colombia: a population-based cohort study. Lancet Reg. Health Am. 12, 100296 (2022).",
"volume": "12",
"year": "2022"
},
{
"DOI": "10.1074/jbc.RA120.016105",
"author": "A Patel",
"doi-asserted-by": "publisher",
"first-page": "17904",
"journal-title": "J. Biol. Chem.",
"key": "2277_CR35",
"unstructured": "Patel, A. et al. Molecular characterization of the RNA-protein complex directing -2/-1 programmed ribosomal frameshifting during arterivirus replicase expression. J. Biol. Chem. 295, 17904–17921 (2020).",
"volume": "295",
"year": "2020"
},
{
"DOI": "10.1093/nar/gkw480",
"author": "S Napthine",
"doi-asserted-by": "publisher",
"first-page": "5491",
"journal-title": "Nucleic Acids Res.",
"key": "2277_CR36",
"unstructured": "Napthine, S. et al. A novel role for poly(C) binding proteins in programmed ribosomal frameshifting. Nucleic Acids Res. 44, 5491–5503 (2016).",
"volume": "44",
"year": "2016"
},
{
"DOI": "10.1038/s41564-020-00846-z",
"author": "N Schmidt",
"doi-asserted-by": "publisher",
"first-page": "339",
"journal-title": "Nat. Microbiol.",
"key": "2277_CR37",
"unstructured": "Schmidt, N. et al. The SARS-CoV-2 RNA-protein interactome in infected human cells. Nat. Microbiol. 6, 339–353 (2021).",
"volume": "6",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2018.12.030",
"author": "X Wang",
"doi-asserted-by": "publisher",
"first-page": "625",
"journal-title": "Cell",
"key": "2277_CR38",
"unstructured": "Wang, X. et al. Regulation of HIV-1 Gag-Pol expression by shiftless, an inhibitor of programmed -1 ribosomal frameshifting. Cell 176, 625–635.e614 (2019).",
"volume": "176",
"year": "2019"
},
{
"DOI": "10.1007/s12250-021-00423-6",
"author": "D Yu",
"doi-asserted-by": "publisher",
"first-page": "1443",
"journal-title": "Virol. Sin.",
"key": "2277_CR39",
"unstructured": "Yu, D. et al. C19orf66 inhibits japanese encephalitis virus replication by targeting -1 PRF and the NS3 protein. Virol. Sin. 36, 1443–1455 (2021).",
"volume": "36",
"year": "2021"
},
{
"DOI": "10.1371/journal.ppat.1005357",
"author": "Y Suzuki",
"doi-asserted-by": "publisher",
"journal-title": "PLoS Pathog.",
"key": "2277_CR40",
"unstructured": "Suzuki, Y. et al. Characterization of RyDEN (C19orf66) as an interferon-stimulated cellular inhibitor against dengue virus replication. PLoS Pathog. 12, e1005357 (2016).",
"volume": "12",
"year": "2016"
},
{
"DOI": "10.1128/JVI.00373-19",
"author": "W Rodriguez",
"doi-asserted-by": "publisher",
"first-page": "e00373",
"journal-title": "J. Virol.",
"key": "2277_CR41",
"unstructured": "Rodriguez, W., Srivastav, K. & Muller, M. C19ORF66 broadly escapes virus-induced endonuclease cleavage and restricts Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 93, e00373–19 (2019).",
"volume": "93",
"year": "2019"
},
{
"DOI": "10.1093/nar/gkad353",
"author": "Y Xu",
"doi-asserted-by": "publisher",
"first-page": "W468",
"journal-title": "Nucleic Acids Res.",
"key": "2277_CR42",
"unstructured": "Xu, Y. et al. PrismNet: predicting protein-RNA interaction using in vivo RNA structural information. Nucleic Acids Res. 51, W468–W477 (2023).",
"volume": "51",
"year": "2023"
},
{
"DOI": "10.7554/eLife.74153",
"author": "JY Lee",
"doi-asserted-by": "publisher",
"journal-title": "Elife",
"key": "2277_CR43",
"unstructured": "Lee, J. Y. et al. Absolute quantitation of individual SARS-CoV-2 RNA molecules provides a new paradigm for infection dynamics and variant sdifferences. Elife 11, e74153 (2022).",
"volume": "11",
"year": "2022"
}
],
"reference-count": 43,
"references-count": 43,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.nature.com/articles/s41392-025-02277-w"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Stem loop binding protein promotes SARS-CoV-2 replication via -1 programmed ribosomal frameshifting",
"type": "journal-article",
"update-policy": "https://doi.org/10.1007/springer_crossmark_policy",
"volume": "10"
}