UNC0638 inhibits SARS-CoV-2 entry by blocking cathepsin L maturation
et al., Journal of Virology, doi:10.1128/jvi.00741-25, Jul 2025
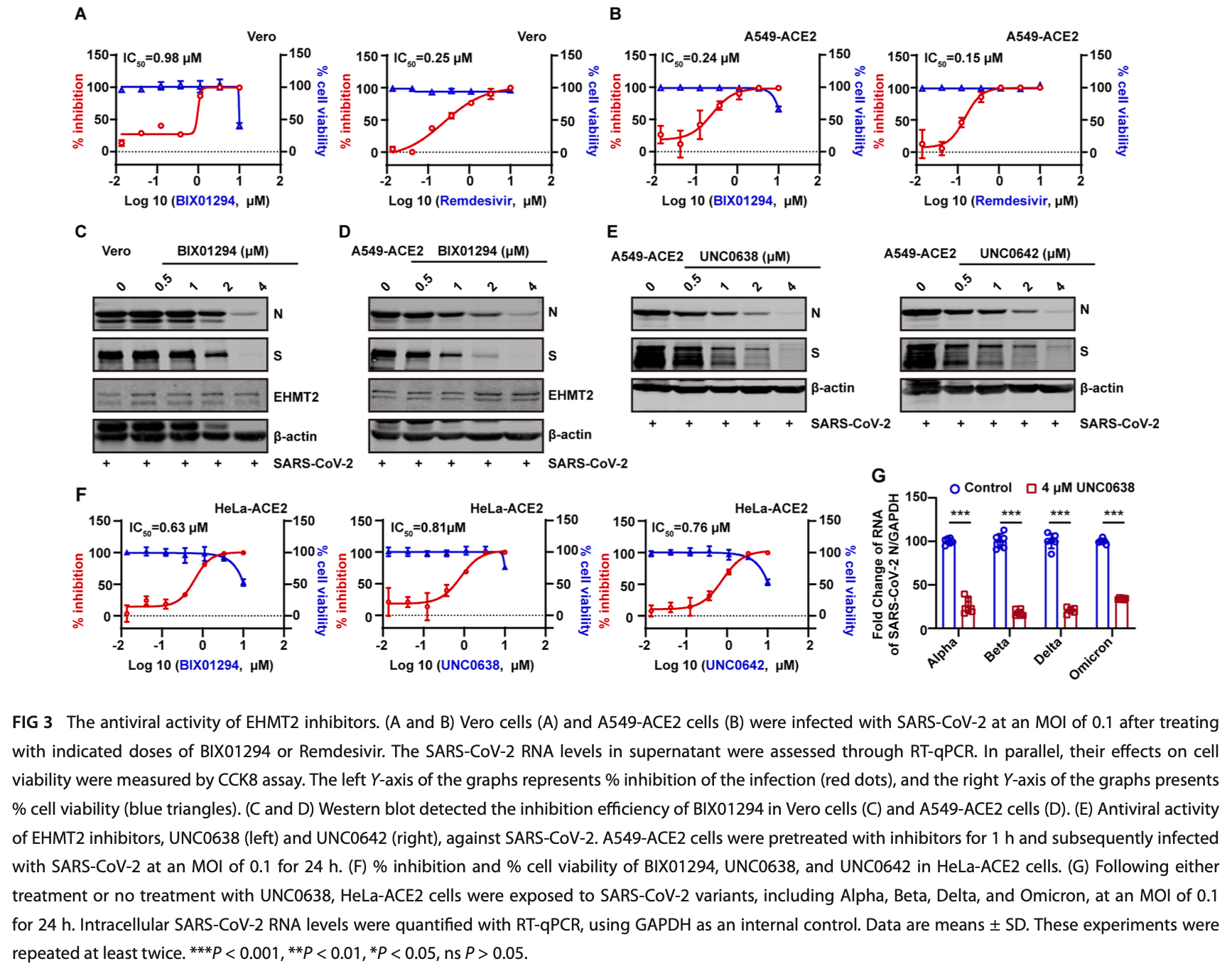
In vitro study showing benefit with UNC0638, an EHMT2 inhibitor, in inhibiting SARS-CoV-2 entry by blocking cathepsin L maturation. Authors identified EHMT2 as a key host factor via ABE-based CRISPR activation screening in A549-ACE2 cells, where EHMT2 inhibition reduced viral infection without enhancing IFN responses to poly(I:C) stimulation.
Chen et al., 22 Jul 2025, multiple countries, peer-reviewed, 12 authors.
Contact: wangjw28@163.com, fyleixb@126.com, wswei@pku.edu.cn.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
UNC0638 inhibits SARS-CoV-2 entry by blocking cathepsin L maturation
Journal of Virology, doi:10.1128/jvi.00741-25
Since the outbreak of SARS-CoV-2, viral mutations have posed significant challenges in identifying therapeutic targets and developing broad-spectrum antiviral drugs. Post-translational modifications of genes involved in interferon production and signaling pathways play a crucial role in regulating interferon responses. In this study, we employed CRISPR-Cas9 screening based on adenine base editors to investigate functional amino acids in 1,278 innate immune-related genes. This approach, which converts A-T base pairs into G-C base pairs to probe the functional importance of specific amino acids, allowed us to identify 17 vital factors involved in SARS-CoV-2 infection. Among the candidate genes, genetic knockdown of EHMT2 exhibited the strongest antiviral effect. Further analysis revealed that UNC0638, a selective inhibitor of EHMT2, significantly reduced the endosomal entry of SARS-CoV-2 in pseudovirus assays. The observed inhibitory effect was consistently observed across multiple SARS-CoV-2 variants, including Alpha, Beta, Delta, and Omicron. Mechanistically, UNC0638 reduced mature cathepsin L (CTSL) levels, impairing the proteolytic cleavage of SARS-CoV-2 spike protein and subsequent membrane fusion, a critical step for viral entry. Our findings uncover EHMT2 as a host dependency factor and reveal the antiviral mecha nism of EHMT2 inhibitors through CTSL maturation blockade. These results advance the understanding of host factors in SARS-CoV-2 infection and provide a strategic framework for developing host-targeted antiviral therapies.
IMPORTANCE In this study, we demonstrated that knockdown or knockout of EHMT2 inhibited SARS-CoV-2 infection, and inhibitors of EHMT2, including UNC0638, UNC0642, and BIX01294 showed similar restrictive effects. Mechanistically, the EHMT2 inhibitor UNC0638 restricts spike-mediated cell entry by inhibiting the maturation of CTSL, a critical protease required for SARS-CoV-2 entry via the endosomal pathway. Importantly, CTSL is not only essential for SARS-CoV-2 but also plays a key role in the entry of other coronaviruses that utilize similar pathways. Therefore, EHMT2 inhibitors could have broader applications as pan-coronavirus therapeutic agents. KEYWORDS SARS-CoV-2, EHMT2, UNC0638, viral entry, cathepsin L C oronaviruses (CoV) are enveloped, positive-sense RNA viruses with genomes of approximately 26-32 kb long with a short untranslated region at both 5′ and 3′ terminal (1-3). The genome encodes four structural proteins: spike (S) protein, envelope protein, nucleocapsid (N) protein, and membrane (M) protein, along with 16 non-structural proteins (NSP1-16) and approximately eight accessory proteins (1-3). Seven coronaviruses are known to infect humans, including severe acute respira tory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), SARS-CoV-2, human coronavirus HKUI (HCoV-HKU1), HCoV-NL63, HCoV-OC43, and HCoV-229E (3). The first three are highly pathogenic and cause..
AUTHOR AFFILIATIONS
References
Baggen, Jacquemyn, Persoons, Vanstreels, Pye et al., TMEM106B is a receptor mediating ACE2-independent SARS-CoV-2 cell entry, Cell, doi:10.1016/j.cell.2023.06.005
Bao, Pan, Xu, Liu, Zhang et al., Unbiased interrogation of functional lysine residues in human proteome, Mol Cell, doi:10.1016/j.molcel.2023.10.033
Bastard, Rosen, Zhang, Michailidis, Hoffmann et al., Autoantibodies against type I IFNs in patients with life-threatening COVID-19, Science
Cao, Gao, Bao, Feng, Mei et al., VV116 versus nirmatrelvir-ritonavir for oral treatment of COVID-19, N Engl J Med, doi:10.1056/NEJMoa2208822
Cao, Wang, Jian, Song, Yisimayi et al., Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies, Nature, doi:10.1038/s41586-021-04385-3
Cuella-Martin, Hayward, Chen, Huang, Taglialatela et al., Functional interrogation of DNA damage response variants with base editing screens, Cell, doi:10.1016/j.molcel.2007.01.017
Drosten, Günther, Preiser, Van Der Werf, Brodt et al., Identification of a novel coronavirus in patients with severe acute respiratory syndrome, N Engl J Med, doi:10.1056/NEJMoa030747
Flury, Breidenbach, Krüger, Voget, Schäkel et al., Cathepsin-targeting SARS-CoV-2 inhibitors: design, synthesis, and biological activity, ACS Pharmacol Transl Sci, doi:10.1021/acsptsci.3c00313
Gaudelli, Komor, Rees, Packer, Badran et al., Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage, Nature, doi:10.1038/nature24644
Guery, Poissy, El Mansouf, Séjourné, Ettahar et al., Clinical features and viral diagnosis of two cases of infection with middle east respiratory syndrome coronavirus: a report of nosocomial transmission, Lancet, doi:10.1016/S0140-6736(13)60982-4
Hadjadj, Yatim, Barnabei, Corneau, Boussier et al., Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients, Science, doi:10.1126/science.abc6027
Hoffmann, Schneider, Rozen-Gagnon, Miles, Schuster et al., TMEM41B is a pan-flavivirus host factor, Cell, doi:10.1016/j.cell.2020.12.005
Huang, Wang, Li, Ren, Zhao et al., Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, Lancet, doi:10.1016/S0140-6736(20)30183-5
Iketani, Liu, Guo, Liu, Chan et al., None, Journal of Virology
Ishiguro, Kitajima, Niinuma, Maruyama, Nishiyama et al., Dual EZH2 and G9a inhibition suppresses multiple myeloma cell proliferation by regulating the interferon signal and IRF4-MYC axis, Cell Death Discov, doi:10.1038/s41420-020-00400-0
Israeli, Finkel, Yahalom-Ronen, Paran, Chitlaru et al., Genome-wide CRISPR screens identify GATA6 as a proviral host factor for SARS-CoV-2 via modulation of ACE2, Nat Commun, doi:10.1038/s41467-022-29896-z
Jiang, Kinch, Brautigam, Chen, Du et al., Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response, Immunity, doi:10.1016/j.immuni.2012.03.022
Kim, Lee, Xiong, Sciaky, Hulbert et al., Targeting the histone methyltransferase G9a activates imprinted genes and improves survival of a mouse model of prader-willi syndrome, Nat Med, doi:10.1038/nm.4257
Koblan, Doman, Wilson, Levy, Tay et al., Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction, Nat Biotechnol, doi:10.1016/j.molcel.2007.01.017
Kubicek, Sullivan, August, Hickey, Zhang et al., Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase, Mol Cell, doi:10.1016/j.molcel.2007.01.017
Kumar, Zhu, Chenna, Hoffpauir, Rademacher et al., Dual inhibitors of SARS-CoV-2 3CL protease and human cathepsin L containing glutamine isosteres are anti-CoV-2 agents, J Am Chem Soc, doi:10.1021/jacs.4c11620
Lei, Dong, Ma, Xiao, Tian et al., Activation and evasion of type I interferon responses by SARS-CoV-2, Nat Commun, doi:10.1038/s41467-020-17665-9
Liu, Qian, Cao, Post-translational modification control of innate immunity, Immunity, doi:10.1016/j.immuni.2016.06.020
Lu, Zhao, Li, Niu, Yang et al., Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding, Lancet, doi:10.1016/S0140-6736(20)30251-8
Ma, Alugubelli, Ma, Vatansever, Scott et al., MPI8 is potent against SARS-CoV-2 by inhibiting dually and selectively the SARS-CoV-2 main protease and the host cathepsin L, ChemMedChem, doi:10.1002/cmdc.202100456
Ma, Zhang, Li, Dong, Wang et al., PLSCR1 suppresses SARS-CoV-2 infection by downregulating cell surface ACE2, J Virol, doi:10.1128/jvi.02085-24
Minkoff, Innate immune evasion strategies of SARS-CoV-2, Nat Rev Microbiol, doi:10.1038/s41579-022-00839-1
Ou, Liu, Lei, Li, Mi et al., Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV, Nat Commun, doi:10.1002/cmdc.202100456
Padmanabhan, Desikan, Dixit, Targeting TMPRSS2 and Cathepsin B/L together may be synergistic against SARS-CoV-2 infection, PLoS Comput Biol, doi:10.1371/journal.pcbi.1008461
Peisley, Wu, Xu, Chen, Hur, Structural basis for ubiquitinmediated antiviral signal activation by RIG-I, Nature, doi:10.1038/nature13140
Prabhakara, Godbole, Sil, Jahnavi, Gulzar et al., Strategies to target SARS-CoV-2 entry and infection using dual mechanisms of inhibition by acidification inhibitors, PLoS Pathog, doi:10.1371/journal.ppat.1009706
Ren, Wang, Wu, Xiang, Guo et al., Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study, Chin Med J, doi:10.1097/CM9.0000000000000722
Sabbatucci, Vitiello, Clemente, Zovi, Boccellino et al., Omicron variant evolution on vaccines and monoclonal antibodies, Inflammopharmacology, doi:10.1007/s10787-023-01253-6
Sakai, Masuda, Tarumoto, Aihara, Tsunoda et al., Genomescale CRISPR-Cas9 screen identifies host factors as potential therapeutic targets for SARS-CoV-2 infection, iScience, doi:10.1002/cmdc.202100456
Singh, Ramĩrez-Carvajal, De, Santos, Golding et al., Inhibition of EHMT2 induces a robust antiviral response against footand-mouth disease and vesicular stomatitis virus infections in bovine cells, J Interferon Cytokine Res, doi:10.1089/jir.2015.0006
Tao, Tzou, Nouhin, Bonilla, Jagannathan et al., SARS-CoV-2 antiviral therapy, Clin Microbiol Rev, doi:10.1128/CMR.00109-21
Wang, Li, Hui, Tiwari, Zhang et al., Cholesterol 25-Hydroxylase inhibits SARS-CoV-2 and other coronaviruses by depleting membrane cholesterol, EMBO J, doi:10.15252/embj.2020106057
Wang, Sheng, Ho, Antibody properties of SARS-CoV-2 Omicron sublineages, Nature, doi:10.1038/s41586-022-04594-4
Wu, Dai, Zhao, Chen, Wang et al., Chloroquine enhances replication of influenza A virus A/WSN/33 (H1N1) in dose-, time-, and MOI-dependent manners in human lung epithelial cells A549, J Med Virol
Xu, Gao, Yang, Jiao, Niu et al., EHMT2 inhibitor BIX-01294 induces endoplasmic reticulum stress mediated apoptosis and autophagy in diffuse large B-cell lymphoma cells, J Cancer, doi:10.7150/jca.48310
Xu, Jiang, Wu, Gaudet, Park et al., PLSCR1 is a cell-autonomous defence factor against SARS-CoV-2 infection, Nature, doi:10.1038/s41586-023-06322-y
Xu, Yue, Protocol for monitoring the endosomal trafficking of membrane proteins in mammalian cells, STAR Protoc, doi:10.1016/j.xpro.2025.103686
Yang, Fan, Xiong, Chen, Zhou et al., Loss of renal tubular G9a benefits acute kidney injury by lowering focal lipid accumulation via CES1, EMBO Rep, doi:10.15252/embr.202256128.32
Yuan, Yin, Meng, Chan, Ye et al., Clofazimine broadly inhibits coronaviruses including SARS-CoV-2, Nature, doi:10.1038/s41586-021-03431-4
Zhang, Shang, Zhang, Liu, Zhao et al., An antibody-based proximity labeling map reveals mechanisms of SARS-CoV-2 inhibition of antiviral immunity, Cell Chem Biol, doi:10.1016/j.chembiol.2021.10.008
Zhang, Sun, Xie, Shang, Wang et al., A viral RNA-dependent RNA polymerase inhibitor VV116 broadly inhibits human coronaviruses and has synergistic potency with 3CLpro inhibitor nirmatrelvir, Sig Transduct Target Ther, doi:10.1038/s41392-023-01587-1
Zhao, Yang, Yang, Zhang, Huang et al., Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development, Signal Transduct Target Ther, doi:10.1038/s41392-021-00558-8
Zhao, Zheng, Chen, Zheng, Li et al., LY6E restricts entry of human coronaviruses, including currently pandemic SARS-CoV-2, J Virol, doi:10.1128/JVI.00562-20
Zhao, Zhong, Zhao, Yong, Tong et al., SARS-CoV-2 spike protein harnesses SNX27-mediated endocytic recycling pathway, MedComm, doi:10.1002/mco2.92
Zhao, Zhu, Zhang, Zhong, Tai et al., Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies, Cell Discov, doi:10.1038/s41421-022-00419-w
Zhou, Zhang, Lei, Xiao, Jiao et al., Sensing of cytoplasmic chromatin by cGAS activates innate immune response in SARS-CoV-2 infection, Signal Transduct Target Ther, doi:10.1002/cmdc.202100456
Zhu, Huang, Sun, Liu, Chen et al., Characteriza tion of ACTN4 as a novel antiviral target against SARS-CoV-2, Signal Transduct Target Ther, doi:10.1038/s41392-024-01956-4
DOI record:
{
"DOI": "10.1128/jvi.00741-25",
"ISSN": [
"0022-538X",
"1098-5514"
],
"URL": "http://dx.doi.org/10.1128/jvi.00741-25",
"abstract": "<jats:title>ABSTRACT</jats:title>\n <jats:sec>\n <jats:title/>\n <jats:p>Since the outbreak of SARS-CoV-2, viral mutations have posed significant challenges in identifying therapeutic targets and developing broad-spectrum antiviral drugs. Post-translational modifications of genes involved in interferon production and signaling pathways play a crucial role in regulating interferon responses. In this study, we employed CRISPR-Cas9 screening based on adenine base editors to investigate functional amino acids in 1,278 innate immune-related genes. This approach, which converts A-T base pairs into G-C base pairs to probe the functional importance of specific amino acids, allowed us to identify 17 vital factors involved in SARS-CoV-2 infection. Among the candidate genes, genetic knockdown of EHMT2 exhibited the strongest antiviral effect. Further analysis revealed that UNC0638, a selective inhibitor of EHMT2, significantly reduced the endosomal entry of SARS-CoV-2 in pseudovirus assays. The observed inhibitory effect was consistently observed across multiple SARS-CoV-2 variants, including Alpha, Beta, Delta, and Omicron. Mechanistically, UNC0638 reduced mature cathepsin L (CTSL) levels, impairing the proteolytic cleavage of SARS-CoV-2 spike protein and subsequent membrane fusion, a critical step for viral entry. Our findings uncover EHMT2 as a host dependency factor and reveal the antiviral mechanism of EHMT2 inhibitors through CTSL maturation blockade. These results advance the understanding of host factors in SARS-CoV-2 infection and provide a strategic framework for developing host-targeted antiviral therapies.</jats:p>\n <jats:sec>\n <jats:title>IMPORTANCE</jats:title>\n <jats:p>In this study, we demonstrated that knockdown or knockout of EHMT2 inhibited SARS-CoV-2 infection, and inhibitors of EHMT2, including UNC0638, UNC0642, and BIX01294 showed similar restrictive effects. Mechanistically, the EHMT2 inhibitor UNC0638 restricts spike-mediated cell entry by inhibiting the maturation of CTSL, a critical protease required for SARS-CoV-2 entry via the endosomal pathway. Importantly, CTSL is not only essential for SARS-CoV-2 but also plays a key role in the entry of other coronaviruses that utilize similar pathways. Therefore, EHMT2 inhibitors could have broader applications as pan-coronavirus therapeutic agents.</jats:p>\n </jats:sec>\n </jats:sec>",
"alternative-id": [
"10.1128/jvi.00741-25"
],
"article-number": "e00741-25",
"assertion": [
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2025-04-25"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "2025-05-25"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 3,
"value": "2025-06-18"
}
],
"author": [
{
"affiliation": [
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/02drdmm93",
"id-type": "ROR"
}
],
"name": "NHC Key Laboratory of System Biology of Pathogens, Christophe Mérieux Laboratory National Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College",
"place": [
"Beijing, China"
]
},
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/00see0y83",
"id-type": "ROR"
}
],
"name": "Key Laboratory of Pathogen Infection Prevention and Control (Peking Union Medical College) Ministry of Education",
"place": [
"Beijing, China"
]
}
],
"family": "Chen",
"given": "Yongjun",
"sequence": "first"
},
{
"affiliation": [
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/02drdmm93",
"id-type": "ROR"
}
],
"name": "NHC Key Laboratory of System Biology of Pathogens, Christophe Mérieux Laboratory National Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College",
"place": [
"Beijing, China"
]
},
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/00see0y83",
"id-type": "ROR"
}
],
"name": "Key Laboratory of Pathogen Infection Prevention and Control (Peking Union Medical College) Ministry of Education",
"place": [
"Beijing, China"
]
}
],
"family": "Shi",
"given": "Yujin",
"sequence": "additional"
},
{
"affiliation": [
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/02drdmm93",
"id-type": "ROR"
}
],
"name": "NHC Key Laboratory of System Biology of Pathogens, Christophe Mérieux Laboratory National Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College",
"place": [
"Beijing, China"
]
},
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/00see0y83",
"id-type": "ROR"
}
],
"name": "Key Laboratory of Pathogen Infection Prevention and Control (Peking Union Medical College) Ministry of Education",
"place": [
"Beijing, China"
]
}
],
"family": "Zuo",
"given": "Xiaoyan",
"sequence": "additional"
},
{
"affiliation": [
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/02drdmm93",
"id-type": "ROR"
}
],
"name": "NHC Key Laboratory of System Biology of Pathogens, Christophe Mérieux Laboratory National Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College",
"place": [
"Beijing, China"
]
},
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/00see0y83",
"id-type": "ROR"
}
],
"name": "Key Laboratory of Pathogen Infection Prevention and Control (Peking Union Medical College) Ministry of Education",
"place": [
"Beijing, China"
]
}
],
"family": "Dong",
"given": "Xiaojing",
"sequence": "additional"
},
{
"affiliation": [
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/02drdmm93",
"id-type": "ROR"
}
],
"name": "NHC Key Laboratory of System Biology of Pathogens, Christophe Mérieux Laboratory National Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College",
"place": [
"Beijing, China"
]
},
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/00see0y83",
"id-type": "ROR"
}
],
"name": "Key Laboratory of Pathogen Infection Prevention and Control (Peking Union Medical College) Ministry of Education",
"place": [
"Beijing, China"
]
}
],
"family": "Xiao",
"given": "Xia",
"sequence": "additional"
},
{
"affiliation": [
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/02drdmm93",
"id-type": "ROR"
}
],
"name": "NHC Key Laboratory of System Biology of Pathogens, Christophe Mérieux Laboratory National Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College",
"place": [
"Beijing, China"
]
},
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/00see0y83",
"id-type": "ROR"
}
],
"name": "Key Laboratory of Pathogen Infection Prevention and Control (Peking Union Medical College) Ministry of Education",
"place": [
"Beijing, China"
]
}
],
"family": "Chen",
"given": "Lan",
"sequence": "additional"
},
{
"affiliation": [
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/02drdmm93",
"id-type": "ROR"
}
],
"name": "NHC Key Laboratory of System Biology of Pathogens, Christophe Mérieux Laboratory National Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College",
"place": [
"Beijing, China"
]
},
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/00see0y83",
"id-type": "ROR"
}
],
"name": "Key Laboratory of Pathogen Infection Prevention and Control (Peking Union Medical College) Ministry of Education",
"place": [
"Beijing, China"
]
}
],
"family": "Xiang",
"given": "Zichun",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-6645-8183",
"affiliation": [
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/02drdmm93",
"id-type": "ROR"
}
],
"name": "NHC Key Laboratory of System Biology of Pathogens, Christophe Mérieux Laboratory National Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College",
"place": [
"Beijing, China"
]
},
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/00see0y83",
"id-type": "ROR"
}
],
"name": "Key Laboratory of Pathogen Infection Prevention and Control (Peking Union Medical College) Ministry of Education",
"place": [
"Beijing, China"
]
}
],
"authenticated-orcid": false,
"family": "Ren",
"given": "Lili",
"sequence": "additional"
},
{
"affiliation": [
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/02szepc22",
"id-type": "ROR"
}
],
"name": "State Key Laboratory of Common Mechanism Research for Major Diseases, Suzhou Institute of Systems Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College",
"place": [
"Suzhou, China"
]
}
],
"family": "Zhou",
"given": "Zhuo",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-8053-2423",
"affiliation": [
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/02v51f717",
"id-type": "ROR"
}
],
"name": "Biomedical Pioneering Innovation Center, Peking-Tsinghua Center for Life Sciences, Peking University Genome Editing Research Center, State Key Laboratory of Gene Function and Modulation Research, School of Life Sciences, Peking University",
"place": [
"Beijing, China"
]
}
],
"authenticated-orcid": false,
"family": "Wei",
"given": "Wensheng",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-3455-6723",
"affiliation": [
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/02drdmm93",
"id-type": "ROR"
}
],
"name": "NHC Key Laboratory of System Biology of Pathogens, Christophe Mérieux Laboratory National Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College",
"place": [
"Beijing, China"
]
},
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/00see0y83",
"id-type": "ROR"
}
],
"name": "Key Laboratory of Pathogen Infection Prevention and Control (Peking Union Medical College) Ministry of Education",
"place": [
"Beijing, China"
]
}
],
"authenticated-orcid": false,
"family": "Lei",
"given": "Xiaobo",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-1116-4559",
"affiliation": [
{
"id": [
{
"asserted-by": "publisher",
"id": "https://ror.org/02drdmm93",
"id-type": "ROR"
}
],
"name": "NHC Key Laboratory of System Biology of Pathogens, Christophe Mérieux Laboratory National Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College",
"place": [
"Beijing, China"
]
}
],
"authenticated-orcid": true,
"family": "Wang",
"given": "Jianwei",
"sequence": "additional"
}
],
"container-title": "Journal of Virology",
"container-title-short": "J Virol",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"journals.asm.org"
]
},
"created": {
"date-parts": [
[
2025,
6,
18
]
],
"date-time": "2025-06-18T09:00:28Z",
"timestamp": 1750237228000
},
"deposited": {
"date-parts": [
[
2025,
7,
22
]
],
"date-time": "2025-07-22T13:05:33Z",
"timestamp": 1753189533000
},
"editor": [
{
"affiliation": [],
"family": "Liu",
"given": "Shan-Lu",
"sequence": "additional"
}
],
"indexed": {
"date-parts": [
[
2025,
7,
23
]
],
"date-time": "2025-07-23T00:04:58Z",
"timestamp": 1753229098506,
"version": "3.41.2"
},
"is-referenced-by-count": 0,
"issue": "7",
"issued": {
"date-parts": [
[
2025,
7,
22
]
]
},
"journal-issue": {
"issue": "7",
"published-print": {
"date-parts": [
[
2025,
7,
22
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
7,
22
]
],
"date-time": "2025-07-22T00:00:00Z",
"timestamp": 1753142400000
}
},
{
"URL": "https://journals.asm.org/non-commercial-tdm-license",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
7,
22
]
],
"date-time": "2025-07-22T00:00:00Z",
"timestamp": 1753142400000
}
}
],
"link": [
{
"URL": "https://journals.asm.org/doi/pdf/10.1128/jvi.00741-25",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://journals.asm.org/doi/pdf/10.1128/jvi.00741-25",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "235",
"original-title": [],
"prefix": "10.1128",
"published": {
"date-parts": [
[
2025,
7,
22
]
]
},
"published-print": {
"date-parts": [
[
2025,
7,
22
]
]
},
"publisher": "American Society for Microbiology",
"reference": [
{
"DOI": "10.1097/CM9.0000000000000722",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_2_2"
},
{
"DOI": "10.1016/S0140-6736(20)30251-8",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_3_2"
},
{
"DOI": "10.1038/s41564-020-0695-z",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_4_2"
},
{
"DOI": "10.1056/NEJMoa030747",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_5_2"
},
{
"DOI": "10.1016/S0140-6736(13)60982-4",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_6_2"
},
{
"DOI": "10.1016/S0140-6736(20)30183-5",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_7_2"
},
{
"DOI": "10.1038/s41586-022-04594-4",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_8_2"
},
{
"DOI": "10.1038/s41586-021-04385-3",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_9_2"
},
{
"DOI": "10.1007/s10787-023-01253-6",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_10_2"
},
{
"DOI": "10.1038/s41392-024-01956-4",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_11_2"
},
{
"DOI": "10.1038/s41392-023-01587-1",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_12_2"
},
{
"DOI": "10.1056/NEJMoa2208822",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_13_2"
},
{
"DOI": "10.1128/CMR.00109-21",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_14_2"
},
{
"DOI": "10.1016/j.cell.2020.12.005",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_15_2"
},
{
"DOI": "10.1016/j.cell.2023.06.005",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_16_2"
},
{
"DOI": "10.1002/mco2.92",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_17_2"
},
{
"DOI": "10.1038/s41467-022-29896-z",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_18_2"
},
{
"DOI": "10.1128/JVI.00562-20",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_19_2"
},
{
"DOI": "10.15252/embj.2020106057",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_20_2"
},
{
"DOI": "10.1038/s41586-023-06322-y",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_21_2"
},
{
"DOI": "10.1128/jvi.02085-24",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_22_2"
},
{
"DOI": "10.1038/s41579-022-00839-1",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_23_2"
},
{
"DOI": "10.1016/j.immuni.2016.06.020",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_24_2"
},
{
"DOI": "10.1016/j.immuni.2012.03.022",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_25_2"
},
{
"DOI": "10.1038/nature13140",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_26_2"
},
{
"DOI": "10.1016/j.molcel.2023.10.033",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_27_2"
},
{
"DOI": "10.1038/nature24644",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_28_2"
},
{
"DOI": "10.1016/j.cell.2021.01.041",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_29_2"
},
{
"DOI": "10.1038/nbt.4172",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_30_2"
},
{
"DOI": "10.1016/j.molcel.2007.01.017",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_31_2"
},
{
"DOI": "10.7150/jca.48310",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_32_2"
},
{
"DOI": "10.15252/embr.202256128.32",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_33_2"
},
{
"DOI": "10.1038/s41420-020-00400-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_34_2"
},
{
"DOI": "10.1038/nm.4257",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_35_2"
},
{
"DOI": "10.1089/jir.2015.0006",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_36_2"
},
{
"DOI": "10.1038/s41586-021-03431-4",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_37_2"
},
{
"DOI": "10.1371/journal.ppat.1009706",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_38_2"
},
{
"DOI": "10.1016/j.xpro.2025.103686",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_39_2"
},
{
"DOI": "10.1126/science.abc6027",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_40_2"
},
{
"DOI": "10.1126/science.abd4585",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_41_2"
},
{
"DOI": "10.1038/s41467-020-17665-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_42_2"
},
{
"DOI": "10.1016/j.chembiol.2021.10.008",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_43_2"
},
{
"DOI": "10.1038/s41392-021-00800-3",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_44_2"
},
{
"DOI": "10.1016/j.isci.2024.110475",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_45_2"
},
{
"DOI": "10.1038/s41467-020-15562-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_46_2"
},
{
"DOI": "10.1371/journal.pcbi.1008461",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_47_2"
},
{
"DOI": "10.1038/s41392-021-00558-8",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_48_2"
},
{
"DOI": "10.1021/jacs.4c11620",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_49_2"
},
{
"DOI": "10.1002/cmdc.202100456",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_50_2"
},
{
"DOI": "10.1038/s41421-022-00419-w",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_51_2"
},
{
"DOI": "10.1021/acsptsci.3c00313",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_52_2"
},
{
"DOI": "10.1002/jmv.24135",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_53_2"
}
],
"reference-count": 52,
"references-count": 52,
"relation": {},
"resource": {
"primary": {
"URL": "https://journals.asm.org/doi/10.1128/jvi.00741-25"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "UNC0638 inhibits SARS-CoV-2 entry by blocking cathepsin L maturation",
"type": "journal-article",
"update-policy": "https://doi.org/10.1128/asmj-crossmark-policy-page",
"volume": "99"
}