Structural and functional insights into Ubl domain-mediated regulation of SARS-CoV-2 PLpro
et al., Biology Direct, doi:10.1186/s13062-025-00690-3, Oct 2025
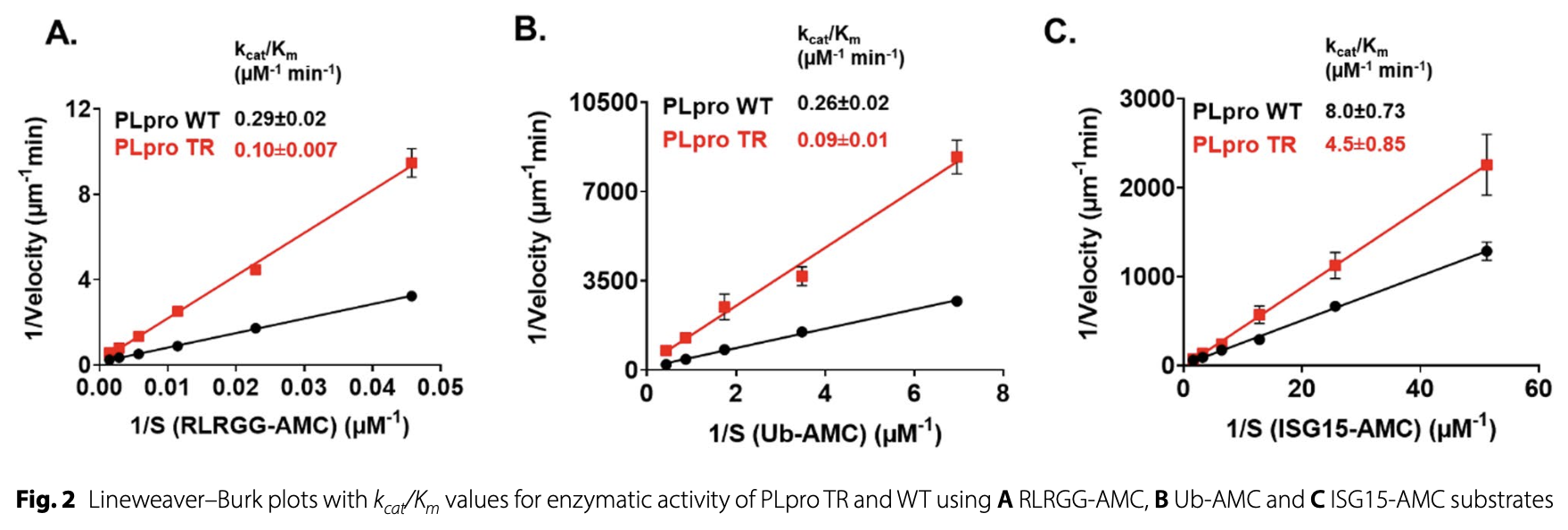
In vitro and in silico study showing structural and functional insights into SARS-CoV-2 PLpro regulation by its ubiquitin-like (Ubl) domain. The study provides mechanistic insights into PLpro regulation that could inform development of allosteric inhibitors targeting sites beyond the active site for COVID-19 treatment.
Arya et al., 3 Oct 2025, peer-reviewed, 4 authors.
Contact: vishalp@barc.gov.in, mukeshk@barc.gov.in.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Structural and functional insights into Ubl domain-mediated regulation of SARS-CoV-2 PLpro
Biology Direct, doi:10.1186/s13062-025-00690-3
domain that hydrolyzes peptide and isopeptide bonds of viral and cellular substrates. PLpro cleaves viral polyproteins to release Nsp1, Nsp2, and Nsp3 which is essential for viral replication [1, 2]. In addition, PLpro removes ubiquitin (Ub) and interferon-stimulated gene product 15 (ISG15) from the cellular protein substrates-a process known as deubiquitination (DUB) and deISGylation, respectively [2-4]. By acting as both a deubiquitinase and a deISGylase, PLpro enables SARS-CoV-2 to evade host immune responses. It impairs the interferon signalling pathway by removing Ub and ISG15 from key signalling proteins, thereby disrupting innate immunity [5-7]. Furthermore, PLpro-mediated deISGylation of the viral nucleocapsid protein enhances viral replication [8].
Supplementary Information The online version contains supplementary material available at h t t p s : / / d o i . o r g / 1 0 . 1 1 8 6 / s 1 3 0 6 2 -0 2 5 -0 0 6 9 0 -3.
Supplementary Material 1. Author contributions R.A.: Planned and performed experiments, analyzed data and written the manuscript, J.G.: Performed experiments, analyzed data and written the manuscript, V.P.: Supervised the research work, analyzed data and written the manuscript, M.K.: Supervised the research work, analyzed data and written the manuscript.
Declarations
Competing interests The authors declare no competing interests.
Publisher's note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Adams, Grosse-Kunstleve, Hung, Ioerger, Mccoy et al., PHENIX: building new software for automated crystallographic structure determination, Acta Crystallogr D Biol Crystallogr, doi:10.1107/S0907444902016657
Al-Homoudi, Engel, Muczynski, Brunzelle, Gavande et al., Human structural homologues of SARS-CoV-2 PLpro as anti-targets: a strategic panel analysis, MicroPubl Biol, doi:10.17912/micropub.biology.001418
Armstrong, Lange, Cesare, Matthews, Nirujogi et al., Biochemical characterization of protease activity of Nsp3 from SARS-CoV-2 and its Inhibition by nanobodies, PLoS ONE, doi:10.1371/journal.pone.0253364
Arya, Kumari, Pandey, Mistry, Bihani et al., Structural insights into SARS-CoV-2 proteins, J Mol Biol, doi:10.1016/j.jmb.2020.11.024
Arya, Prashar, Kumar, Evaluating stability and activity of SARS-CoV-2 PLpro for High-throughput screening of inhibitors, Mol Biotechnol, doi:10.1007/s12033-021-00383-y
Arya, Prashar, Kumar, Identification and characterization of aurintricarboxylic acid as a potential inhibitor of SARS-CoV-2 PLpro, Int J Biol Macromol, doi:10.1016/j.ijbiomac.2023.123347
Berendsen, Van Der Spoel, Van Drunen, A message-passing parallel molecular dynamics implementation, Comput Phys Commun, doi:10.1016/0010-4655(95)00042-E
Báez-Santos, Mielech, Deng, Baker, Mesecar, Catalytic function and substrate specificity of the Papain-like protease domain of nsp3 from the middle East respiratory syndrome coronavirus, J Virol, doi:10.1128/JVI.01294-14
Békés, Van Der Heden Van Noort, Ekkebus, Ovaa, Huang et al., Recognition of Lys48-Linked Di-ubiquitin and deubiquitinating activities of the SARS coronavirus Papain-like protease, Mol Cell, doi:10.1016/j.molcel.2016.04.016
Clasman, Báez-Santos, Mettelman, 'brien, Baker et al., X-ray structure and enzymatic activity profile of a core Papain-like protease of MERS coronavirus with utility for structure-based drug design, Sci Rep, doi:10.1038/srep40292
Clerici, Vargas, Faesen, Sixma, The DUSP-Ubl domain of USP4 enhances its catalytic efficiency by promoting ubiquitin exchange, Nat Commun, doi:10.1038/ncomms6399
Comeau, Gatchell, Vajda, Camacho, ClusPro: an automated Docking and discrimination method for the prediction of protein complexes, Bioinformatics, doi:10.1093/bioinformatics/btg371
Emsley, Lohkamp, Scott, Cowtan, Features and development of coot, Acta Crystallogr D Biol Crystallogr, doi:10.1107/S0907444910007493
Faesen, Vargas, Sixma, The role of UBL domains in ubiquitinspecific proteases, Biochem Soc Trans, doi:10.1042/BST20120004
Ferreira, Pillaiyar, Hirata, Poso, Kronenberger, Inhibitor induced conformational changes in SARS-COV-2 papain-like protease, Sci Rep, doi:10.1038/s41598-022-15181-y
Ferreira, Villanueva, Adem, Fadl, Alzyoud et al., Identification of novel allosteric sites of SARS-CoV-2 papain-like protease (PLpro) for the development of COVID-19 antivirals, J Biol Chem, doi:10.1016/j.jbc.2024.107821
Frieman, Ratia, Johnston, Mesecar, Baric, Severe acute respiratory syndrome coronavirus Papain-like protease Ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-κB signaling, J Virol, doi:10.1128/JVI.02220-08
Hu, Li, Song, Jeffrey, Chenova et al., Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14, EMBO J, doi:10.1038/sj.emboj.7600832
Huang, Wang, Zhong, Zhang, Zhang et al., Molecular architecture of coronavirus double-membrane vesicle pore complex, Nature, doi:10.1038/s41586-024-07817-y
Kabsch, Xds, None, Acta Crystallogr D Biol Crystallogr, doi:10.1107/S0907444909047337
Klemm, Ebert, Calleja, Allison, Richardson et al., Mechanism and inhibition of the papain-like protease, plpro, of SARS-CoV-2, EMBO J, doi:10.15252/embj.2020106275
Komander, Clague, Urbé, Breaking the chains: structure and function of the deubiquitinases, Nat Rev Mol Cell Biol, doi:10.1038/nrm2731
Kozakov, Hall, Xia, Porter, Padhorny et al., The cluspro web server for protein-protein Docking, Nat Protoc, doi:10.1038/nprot.2016.169
Lee, Lei, Santarsiero, Gatuz, Cao et al., Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV, ACS Chem Biol, doi:10.1021/cb500917m
Liu, Lee, Parker, Acharya, Chiang et al., ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity, Nat Microbiol, doi:10.1038/s41564-021-00884-1
Lu, Zhao, Yu, Kang, Yang, Targeting ubiquitin-specific protease 7 (USP7) in cancer: a new insight to overcome drug resistance, Front Pharmacol, doi:10.3389/fphar.2021.648491
Mccoy, Grosse-Kunstleve, Adams, Winn, Storoni et al., Phaser crystallographic software, J Appl Crystallogr, doi:10.1107/S0021889807021206
Munnur, Teo, Eggermont, Lee, Thery et al., Altered isgylation drives aberrant macrophagedependent immune responses during SARS-CoV-2 infection, Nat Immunol, doi:10.1038/s41590-021-01035-8
Osipiuk, Azizi, Dvorkin, Endres, Jedrzejczak et al., Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors, Nat Commun, doi:10.1038/s41467-021-21060-3
Patchett, Lv, Rut, Békés, Drag et al., A molecular sensor determines the ubiquitin substrate specificity of SARS-CoV-2 papainlike protease, Cell Rep, doi:10.1016/j.celrep.2021.109754
Peth, Besche, Goldberg, Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6 which causes 20s gate opening, Mol Cell, doi:10.1016/j.molcel.2009.11.015
Ran, Zhu, Chen, Ni, Mu, Papain-like protease of SARS-CoV-2 inhibits RLR signaling in a deubiquitination-dependent and deubiquitination-independent manner, Front Immunol, doi:10.3389/fimmu.2022.947272
Rehman, Armstrong, Lange, Kristariyanto, Gräwert et al., Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2, Mol Cell, doi:10.1016/j.molcel.2021.08.024
Rhamadianti, Abe, Tanaka, Ono, Katayama et al., SARS-CoV-2 papain-like protease inhibits isgylation of the viral nucleocapsid protein to evade host anti-viral immunity, J Virol. n.d, doi:10.1128/jvi.00855-24
Shin, Mukherjee, Grewe, Bojkova, Baek et al., Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity, Nature, doi:10.1038/s41586-020-2601-5
Srinivasan, Brognaro, Prabhu, De Souza, Günther et al., Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papainlike protease, Commun Biol, doi:10.1038/s42003-022-03737-7
Sulea, Lindner, Purisima, Ménard, Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus Papain-like protease?, J Virol, doi:10.1128/jvi.79.7.4550-4551.2005
Vliet, Huynh, Palà, Patel, Singer et al., Ubiquitin variants potently inhibit SARS-CoV-2 PLpro and viral replication via a novel site distal to the protease active site, PLoS Pathog, doi:10.1371/journal.ppat.1011065
Wydorski, Osipiuk, Lanham, Tesar, Endres et al., Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin, Nat Commun, doi:10.1038/s41467-023-38031-5
DOI record:
{
"DOI": "10.1186/s13062-025-00690-3",
"ISSN": [
"1745-6150"
],
"URL": "http://dx.doi.org/10.1186/s13062-025-00690-3",
"alternative-id": [
"690"
],
"article-number": "102",
"assertion": [
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Received",
"name": "received",
"order": 1,
"value": "20 June 2025"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "3 September 2025"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "First Online",
"name": "first_online",
"order": 3,
"value": "3 October 2025"
},
{
"group": {
"label": "Declarations",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 1
},
{
"group": {
"label": "Competing interests",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 2,
"value": "The authors declare no competing interests."
}
],
"author": [
{
"affiliation": [],
"family": "Arya",
"given": "Rimanshee",
"sequence": "first"
},
{
"affiliation": [],
"family": "Ganesh",
"given": "Janani",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Prashar",
"given": "Vishal",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Kumar",
"given": "Mukesh",
"sequence": "additional"
}
],
"container-title": "Biology Direct",
"container-title-short": "Biol Direct",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"link.springer.com"
]
},
"created": {
"date-parts": [
[
2025,
10,
3
]
],
"date-time": "2025-10-03T11:30:33Z",
"timestamp": 1759491033000
},
"deposited": {
"date-parts": [
[
2025,
10,
3
]
],
"date-time": "2025-10-03T11:30:34Z",
"timestamp": 1759491034000
},
"funder": [
{
"DOI": "10.13039/501100005031",
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100005031",
"id-type": "DOI"
}
],
"name": "Bhabha Atomic Research Centre"
}
],
"indexed": {
"date-parts": [
[
2025,
10,
7
]
],
"date-time": "2025-10-07T01:13:45Z",
"timestamp": 1759799625631,
"version": "build-2065373602"
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2025,
10,
3
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2025,
12
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by-nc-nd/4.0",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
10,
3
]
],
"date-time": "2025-10-03T00:00:00Z",
"timestamp": 1759449600000
}
},
{
"URL": "https://creativecommons.org/licenses/by-nc-nd/4.0",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
10,
3
]
],
"date-time": "2025-10-03T00:00:00Z",
"timestamp": 1759449600000
}
}
],
"link": [
{
"URL": "https://link.springer.com/content/pdf/10.1186/s13062-025-00690-3.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://link.springer.com/article/10.1186/s13062-025-00690-3/fulltext.html",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://link.springer.com/content/pdf/10.1186/s13062-025-00690-3.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"prefix": "10.1186",
"published": {
"date-parts": [
[
2025,
10,
3
]
]
},
"published-online": {
"date-parts": [
[
2025,
10,
3
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference": [
{
"DOI": "10.1016/j.jmb.2020.11.024",
"author": "R Arya",
"doi-asserted-by": "publisher",
"first-page": "166725",
"journal-title": "J Mol Biol",
"key": "690_CR1",
"unstructured": "Arya R, Kumari S, Pandey B, Mistry H, Bihani SC, Das A, Prashar V, Gupta GD, Panicker L, Kumar M. Structural insights into SARS-CoV-2 proteins. J Mol Biol. 2021;433:166725. https://doi.org/10.1016/j.jmb.2020.11.024.",
"volume": "433",
"year": "2021"
},
{
"DOI": "10.1371/journal.pone.0253364",
"author": "LA Armstrong",
"doi-asserted-by": "publisher",
"first-page": "e0253364",
"journal-title": "PLoS ONE",
"key": "690_CR2",
"unstructured": "Armstrong LA, Lange SM, Dee Cesare V, Matthews SP, Nirujogi RS, Cole I, Hope A, Cunningham F, Toth R, Mukherjee R, Bojkova D, Gruber F, Gray D, Wyatt PG, Cinatl J, Dikic I, Davies P, Kulathu Y. Biochemical characterization of protease activity of Nsp3 from SARS-CoV-2 and its Inhibition by nanobodies. PLoS ONE. 2021;16:e0253364. https://doi.org/10.1371/journal.pone.0253364.",
"volume": "16",
"year": "2021"
},
{
"DOI": "10.1128/jvi.79.7.4550-4551.2005",
"author": "T Sulea",
"doi-asserted-by": "publisher",
"first-page": "4550",
"journal-title": "J Virol",
"key": "690_CR3",
"unstructured": "Sulea T, Lindner HA, Purisima EO, Ménard R. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus Papain-like protease? J Virol. 2005;79:4550–1. https://doi.org/10.1128/jvi.79.7.4550-4551.2005.",
"volume": "79",
"year": "2005"
},
{
"DOI": "10.1038/s41467-023-38031-5",
"author": "PM Wydorski",
"doi-asserted-by": "publisher",
"first-page": "2366",
"journal-title": "Nat Commun",
"key": "690_CR4",
"unstructured": "Wydorski PM, Osipiuk J, Lanham BT, Tesar C, Endres M, Engle E, Jedrzejczak R, Mullapudi V, Michalska K, Fidelis K, Fushman D, Joachimiak A, Joachimiak LA. Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin. Nat Commun. 2023;14:2366. https://doi.org/10.1038/s41467-023-38031-5.",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.3389/fimmu.2022.947272",
"doi-asserted-by": "publisher",
"key": "690_CR5",
"unstructured": "Ran X-H, Zhu J-W, Chen Y-Y, Ni R-Z, Mu D. Papain-like protease of SARS-CoV-2 inhibits RLR signaling in a deubiquitination-dependent and deubiquitination-independent manner. Front Immunol. 2022;13. https://doi.org/10.3389/fimmu.2022.947272."
},
{
"DOI": "10.1038/s41564-021-00884-1",
"author": "G Liu",
"doi-asserted-by": "publisher",
"first-page": "467",
"journal-title": "Nat Microbiol",
"key": "690_CR6",
"unstructured": "Liu G, Lee J-H, Parker ZM, Acharya D, Chiang JJ, van Gent M, Riedl W, Davis-Gardner ME, Wies E, Chiang C, Gack MU. ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat Microbiol. 2021;6:467–78. https://doi.org/10.1038/s41564-021-00884-1.",
"volume": "6",
"year": "2021"
},
{
"DOI": "10.1038/s41590-021-01035-8",
"author": "D Munnur",
"doi-asserted-by": "publisher",
"first-page": "1416",
"journal-title": "Nat Immunol",
"key": "690_CR7",
"unstructured": "Munnur D, Teo Q, Eggermont D, Lee HHY, Thery F, Ho J, van Leur SW, Ng WWS, Siu LYL, Beling A, Ploegh H, Pinto-Fernandez A, Damianou A, Kessler B, Impens F, Mok CKP, Sanyal S. Altered isgylation drives aberrant macrophage-dependent immune responses during SARS-CoV-2 infection. Nat Immunol. 2021;22:1416–27. https://doi.org/10.1038/s41590-021-01035-8.",
"volume": "22",
"year": "2021"
},
{
"DOI": "10.1128/jvi.00855-24",
"doi-asserted-by": "publisher",
"key": "690_CR8",
"unstructured": "Rhamadianti AF, Abe T, Tanaka T, Ono C, Katayama H, Makino Y, Deng L, Matsui C, Moriishi K, Shima F, Matsuura Y, Shoji I. SARS-CoV-2 papain-like protease inhibits isgylation of the viral nucleocapsid protein to evade host anti-viral immunity. J Virol. n.d.;98(e00855–24). https://doi.org/10.1128/jvi.00855-24."
},
{
"DOI": "10.1038/s41586-024-07817-y",
"author": "Y Huang",
"doi-asserted-by": "publisher",
"first-page": "224",
"journal-title": "Nature",
"key": "690_CR9",
"unstructured": "Huang Y, Wang T, Zhong L, Zhang W, Zhang Y, Yu X, Yuan S, Ni T. Molecular architecture of coronavirus double-membrane vesicle pore complex. Nature. 2024;633:224–31. https://doi.org/10.1038/s41586-024-07817-y.",
"volume": "633",
"year": "2024"
},
{
"DOI": "10.1038/s41586-020-2601-5",
"author": "D Shin",
"doi-asserted-by": "publisher",
"first-page": "657",
"journal-title": "Nature",
"key": "690_CR10",
"unstructured": "Shin D, Mukherjee R, Grewe D, Bojkova D, Baek K, Bhattacharya A, Schulz L, Widera M, Mehdipour AR, Tascher G, Geurink PP, Wilhelm A, van der Heden van Noort GJ, Ovaa H, Müller S, Knobeloch K-P, Rajalingam K, Schulman BA, Cinatl J, Hummer G, Ciesek S, Dikic I. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–62. https://doi.org/10.1038/s41586-020-2601-5.",
"volume": "587",
"year": "2020"
},
{
"DOI": "10.1016/j.molcel.2016.04.016",
"author": "M Békés",
"doi-asserted-by": "publisher",
"first-page": "572",
"journal-title": "Mol Cell",
"key": "690_CR11",
"unstructured": "Békés M, van der Heden van Noort GJ, Ekkebus R, Ovaa H, Huang TT, Lima CD. Recognition of Lys48-Linked Di-ubiquitin and deubiquitinating activities of the SARS coronavirus Papain-like protease. Mol Cell. 2016;62:572–85. https://doi.org/10.1016/j.molcel.2016.04.016.",
"volume": "62",
"year": "2016"
},
{
"DOI": "10.15252/embj.2020106275",
"author": "T Klemm",
"doi-asserted-by": "publisher",
"first-page": "e106275",
"journal-title": "EMBO J",
"key": "690_CR12",
"unstructured": "Klemm T, Ebert G, Calleja DJ, Allison CC, Richardson LW, Bernardini JP, Lu BG, Kuchel NW, Grohmann C, Shibata Y, Gan ZY, Cooney JP, Doerflinger M, Au AE, Blackmore TR, van der Heden van Noort PP, Geurink GJ, Ovaa H, Newman J, Riboldi-Tunnicliffe A, Czabotar PE, Mitchell JP, Feltham R, Lechtenberg BC, Lowes KN, Dewson G, Pellegrini M, Lessene G, Komander D. Mechanism and inhibition of the papain‐like protease, plpro, of SARS‐CoV‐2. EMBO J. 2020;39:e106275. https://doi.org/10.15252/embj.2020106275.",
"volume": "39",
"year": "2020"
},
{
"DOI": "10.1038/s42003-022-03737-7",
"author": "V Srinivasan",
"doi-asserted-by": "publisher",
"first-page": "805",
"journal-title": "Commun Biol",
"key": "690_CR13",
"unstructured": "Srinivasan V, Brognaro H, Prabhu PR, de Souza EE, Günther S, Reinke PYA, Lane TJ, Ginn H, Han H, Ewert W, Sprenger J, Koua FHM, Falke S, Werner N, Andaleeb H, Ullah N, Franca BA, Wang M, Barra ALC, Perbandt M, Schwinzer M, Schmidt C, Brings L, Lorenzen K, Schubert R, Machado RRG, Candido ED, Oliveira DBL, Durigon EL, Niebling S, Garcia AS, Yefanov O, Lieske J, Gelisio L, Domaracky M, Middendorf P, Groessler M, Trost F, Galchenkova M, Mashhour AR, Saouane S, Hakanpää J, Wolf M, Alai MG, Turk D, Pearson AR, Chapman HN, Hinrichs W, Wrenger C, Meents A, Betzel C. Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease. Commun Biol. 2022;5:805. https://doi.org/10.1038/s42003-022-03737-7.",
"volume": "5",
"year": "2022"
},
{
"DOI": "10.1371/journal.ppat.1011065",
"author": "VJE Van Vliet",
"doi-asserted-by": "publisher",
"first-page": "e1011065",
"journal-title": "PLoS Pathog",
"key": "690_CR14",
"unstructured": "Van Vliet VJE, Huynh N, Palà J, Patel A, Singer A, Slater C, Chung J, van Huizen M, Teyra J, Miersch S, Luu G-K, Ye W, Sharma N, Ganaie SS, Russell R, Chen C, Maynard M, Amarasinghe GK, Mark BL, Kikkert M, Sidhu SS. Ubiquitin variants potently inhibit SARS-CoV-2 PLpro and viral replication via a novel site distal to the protease active site. PLoS Pathog. 2022;18:e1011065. https://doi.org/10.1371/journal.ppat.1011065.",
"volume": "18",
"year": "2022"
},
{
"DOI": "10.1128/JVI.01294-14",
"author": "YM Báez-Santos",
"doi-asserted-by": "publisher",
"first-page": "12511",
"journal-title": "J Virol",
"key": "690_CR15",
"unstructured": "Báez-Santos YM, Mielech AM, Deng X, Baker S, Mesecar AD. Catalytic function and substrate specificity of the Papain-like protease domain of nsp3 from the middle East respiratory syndrome coronavirus. J Virol. 2014;88:12511–27. https://doi.org/10.1128/JVI.01294-14.",
"volume": "88",
"year": "2014"
},
{
"DOI": "10.1038/srep40292",
"author": "JR Clasman",
"doi-asserted-by": "publisher",
"first-page": "40292",
"journal-title": "Sci Rep",
"key": "690_CR16",
"unstructured": "Clasman JR, Báez-Santos YM, Mettelman RC, O’Brien A, Baker SC, Mesecar AD. X-ray structure and enzymatic activity profile of a core Papain-like protease of MERS coronavirus with utility for structure-based drug design. Sci Rep. 2017;7:40292. https://doi.org/10.1038/srep40292.",
"volume": "7",
"year": "2017"
},
{
"DOI": "10.1128/JVI.02220-08",
"author": "M Frieman",
"doi-asserted-by": "publisher",
"first-page": "6689",
"journal-title": "J Virol",
"key": "690_CR17",
"unstructured": "Frieman M, Ratia K, Johnston RE, Mesecar AD, Baric RS. Severe acute respiratory syndrome coronavirus Papain-like protease Ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-κB signaling. J Virol. 2009;83:6689–705. https://doi.org/10.1128/JVI.02220-08.",
"volume": "83",
"year": "2009"
},
{
"DOI": "10.1042/BST20120004",
"author": "AC Faesen",
"doi-asserted-by": "publisher",
"first-page": "539",
"journal-title": "Biochem Soc Trans",
"key": "690_CR18",
"unstructured": "Faesen AC, Luna-Vargas MPA, Sixma TK. The role of UBL domains in ubiquitin-specific proteases. Biochem Soc Trans. 2012;40:539–45. https://doi.org/10.1042/BST20120004.",
"volume": "40",
"year": "2012"
},
{
"DOI": "10.1038/nrm2731",
"author": "D Komander",
"doi-asserted-by": "publisher",
"first-page": "550",
"journal-title": "Nat Rev Mol Cell Biol",
"key": "690_CR19",
"unstructured": "Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–63. https://doi.org/10.1038/nrm2731.",
"volume": "10",
"year": "2009"
},
{
"DOI": "10.1038/sj.emboj.7600832",
"author": "M Hu",
"doi-asserted-by": "publisher",
"first-page": "3747",
"journal-title": "EMBO J",
"key": "690_CR20",
"unstructured": "Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, Cohen RE, Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24:3747–56. https://doi.org/10.1038/sj.emboj.7600832.",
"volume": "24",
"year": "2005"
},
{
"DOI": "10.1016/j.molcel.2009.11.015",
"author": "A Peth",
"doi-asserted-by": "publisher",
"first-page": "794",
"journal-title": "Mol Cell",
"key": "690_CR21",
"unstructured": "Peth A, Besche HC, Goldberg AL. Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6 which causes 20s gate opening. Mol Cell. 2009;36:794–804. https://doi.org/10.1016/j.molcel.2009.11.015.",
"volume": "36",
"year": "2009"
},
{
"DOI": "10.1038/ncomms6399",
"author": "M Clerici",
"doi-asserted-by": "publisher",
"first-page": "5399",
"journal-title": "Nat Commun",
"key": "690_CR22",
"unstructured": "Clerici M, Luna-Vargas MPA, Faesen AC, Sixma TK. The DUSP–Ubl domain of USP4 enhances its catalytic efficiency by promoting ubiquitin exchange. Nat Commun. 2014;5:5399. https://doi.org/10.1038/ncomms6399.",
"volume": "5",
"year": "2014"
},
{
"DOI": "10.3389/fphar.2021.648491",
"author": "J Lu",
"doi-asserted-by": "publisher",
"first-page": "648491",
"journal-title": "Front Pharmacol",
"key": "690_CR23",
"unstructured": "Lu J, Zhao H, Yu C, Kang Y, Yang X. Targeting ubiquitin-specific protease 7 (USP7) in cancer: a new insight to overcome drug resistance. Front Pharmacol. 2021;12:648491. https://doi.org/10.3389/fphar.2021.648491.",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1007/s12033-021-00383-y",
"author": "R Arya",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Mol Biotechnol",
"key": "690_CR24",
"unstructured": "Arya R, Prashar V, Kumar M. Evaluating stability and activity of SARS-CoV-2 PLpro for High-throughput screening of inhibitors. Mol Biotechnol. 2022;64:1–8. https://doi.org/10.1007/s12033-021-00383-y.",
"volume": "64",
"year": "2022"
},
{
"DOI": "10.1016/j.ijbiomac.2023.123347",
"author": "R Arya",
"doi-asserted-by": "publisher",
"first-page": "123347",
"journal-title": "Int J Biol Macromol",
"key": "690_CR25",
"unstructured": "Arya R, Prashar V, Kumar M. Identification and characterization of aurintricarboxylic acid as a potential inhibitor of SARS-CoV-2 PLpro. Int J Biol Macromol. 2023;252:123347. https://doi.org/10.1016/j.ijbiomac.2023.123347.",
"volume": "252",
"year": "2023"
},
{
"DOI": "10.1107/S0907444909047337",
"author": "W Kabsch",
"doi-asserted-by": "publisher",
"first-page": "125",
"journal-title": "Acta Crystallogr D Biol Crystallogr",
"key": "690_CR26",
"unstructured": "Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. https://doi.org/10.1107/S0907444909047337.",
"volume": "66",
"year": "2010"
},
{
"DOI": "10.1107/S0021889807021206",
"author": "AJ McCoy",
"doi-asserted-by": "publisher",
"first-page": "658",
"journal-title": "J Appl Crystallogr",
"key": "690_CR27",
"unstructured": "McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–74. https://doi.org/10.1107/S0021889807021206.",
"volume": "40",
"year": "2007"
},
{
"DOI": "10.1107/S0907444902016657",
"author": "PD Adams",
"doi-asserted-by": "publisher",
"first-page": "1948",
"journal-title": "Acta Crystallogr D Biol Crystallogr",
"key": "690_CR28",
"unstructured": "Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. https://doi.org/10.1107/S0907444902016657.",
"volume": "58",
"year": "2002"
},
{
"DOI": "10.1107/S0907444910007493",
"author": "P Emsley",
"doi-asserted-by": "publisher",
"first-page": "486",
"journal-title": "Acta Crystallogr D Biol Crystallogr",
"key": "690_CR29",
"unstructured": "Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. https://doi.org/10.1107/S0907444910007493.",
"volume": "66",
"year": "2010"
},
{
"DOI": "10.1093/bioinformatics/btg371",
"author": "SR Comeau",
"doi-asserted-by": "publisher",
"first-page": "45",
"journal-title": "Bioinformatics",
"key": "690_CR30",
"unstructured": "Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: an automated Docking and discrimination method for the prediction of protein complexes. Bioinformatics. 2004;20:45–50. https://doi.org/10.1093/bioinformatics/btg371.",
"volume": "20",
"year": "2004"
},
{
"DOI": "10.1038/nprot.2016.169",
"author": "D Kozakov",
"doi-asserted-by": "publisher",
"first-page": "255",
"journal-title": "Nat Protoc",
"key": "690_CR31",
"unstructured": "Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, Beglov D, Vajda S. The cluspro web server for protein–protein Docking. Nat Protoc. 2017;12:255–78. https://doi.org/10.1038/nprot.2016.169.",
"volume": "12",
"year": "2017"
},
{
"DOI": "10.1016/0010-4655(95)00042-E",
"author": "HJC Berendsen",
"doi-asserted-by": "publisher",
"first-page": "43",
"journal-title": "Comput Phys Commun",
"key": "690_CR32",
"unstructured": "Berendsen HJC, van der Spoel D, van Drunen R. A message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91:43–56. https://doi.org/10.1016/0010-4655(95)00042-E.",
"volume": "91",
"year": "1995"
},
{
"DOI": "10.1016/j.jbc.2024.107821",
"author": "JC Ferreira",
"doi-asserted-by": "publisher",
"first-page": "107821",
"journal-title": "J Biol Chem",
"key": "690_CR33",
"unstructured": "Ferreira JC, Villanueva AJ, Al Adem K, Fadl S, Alzyoud L, Ghattas MA, Rabeh WM. Identification of novel allosteric sites of SARS-CoV-2 papain-like protease (PLpro) for the development of COVID-19 antivirals. J Biol Chem. 2024;300:107821. https://doi.org/10.1016/j.jbc.2024.107821.",
"volume": "300",
"year": "2024"
},
{
"DOI": "10.1038/s41467-021-21060-3",
"author": "J Osipiuk",
"doi-asserted-by": "publisher",
"first-page": "743",
"journal-title": "Nat Commun",
"key": "690_CR34",
"unstructured": "Osipiuk J, Azizi S-A, Dvorkin S, Endres M, Jedrzejczak R, Jones KA, Kang S, Kathayat RS, Kim Y, Lisnyak VG, Maki SL, Nicolaescu V, Taylor CA, Tesar C, Zhang Y-A, Zhou Z, Randall G, Michalska K, Snyder SA, Dickinson BC, Joachimiak A. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat Commun. 2021;12:743. https://doi.org/10.1038/s41467-021-21060-3.",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.17912/micropub.biology.001418",
"author": "AI Al-Homoudi",
"doi-asserted-by": "publisher",
"journal-title": "MicroPubl Biol",
"key": "690_CR35",
"unstructured": "Al-Homoudi AI, Engel J, Muczynski MD, Brunzelle JS, Gavande NS, Kovari LC. Human structural homologues of SARS-CoV-2 PLpro as anti-targets: a strategic panel analysis. MicroPubl Biol. 2025. https://doi.org/10.17912/micropub.biology.001418.",
"year": "2025"
},
{
"DOI": "10.1021/cb500917m",
"author": "H Lee",
"doi-asserted-by": "publisher",
"first-page": "1456",
"issue": "6",
"journal-title": "ACS Chem Biol",
"key": "690_CR36",
"unstructured": "Lee H, Lei H, Santarsiero BD, Gatuz JL, Cao S, Rice AJ, Patel K, Szypulinski MZ, Ojeda I, Ghosh AK, Johnson ME. Inhibitor recognition specificity of MERS–CoV papain–like protease may differ from that of SARS–CoV. ACS Chem Biol. 2015;10(6):1456–65. https://doi.org/10.1021/cb500917m.",
"volume": "10",
"year": "2015"
},
{
"DOI": "10.1038/s41598-022-15181-y",
"author": "GM Ferreira",
"doi-asserted-by": "publisher",
"first-page": "11585",
"journal-title": "Sci Rep",
"key": "690_CR37",
"unstructured": "Ferreira GM, Pillaiyar T, Hirata MH, Poso A, Kronenberger T. Inhibitor induced conformational changes in SARS-COV-2 papain-like protease. Sci Rep. 2022;12:11585. https://doi.org/10.1038/s41598-022-15181-y.",
"volume": "12",
"year": "2022"
},
{
"DOI": "10.1016/j.molcel.2021.08.024",
"author": "SA Abdul Rehman",
"doi-asserted-by": "publisher",
"first-page": "4176",
"journal-title": "Mol Cell",
"key": "690_CR38",
"unstructured": "Abdul Rehman SA, Armstrong LA, Lange SM, Kristariyanto YA, Gräwert TW, Knebel A, Svergun DI, Kulathu Y. Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2. Mol Cell. 2021;81:4176–e41906. https://doi.org/10.1016/j.molcel.2021.08.024.",
"volume": "81",
"year": "2021"
},
{
"DOI": "10.1016/j.celrep.2021.109754",
"doi-asserted-by": "publisher",
"key": "690_CR39",
"unstructured": "Patchett S, Lv Z, Rut W, Békés M, Drag M, Olsen SK, Huang TT. A molecular sensor determines the ubiquitin substrate specificity of SARS-CoV-2 papain-like protease. Cell Rep. 2021;36. https://doi.org/10.1016/j.celrep.2021.109754."
}
],
"reference-count": 39,
"references-count": 39,
"relation": {
"has-preprint": [
{
"asserted-by": "object",
"id": "10.21203/rs.3.rs-6938209/v1",
"id-type": "doi"
}
]
},
"resource": {
"primary": {
"URL": "https://biologydirect.biomedcentral.com/articles/10.1186/s13062-025-00690-3"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Structural and functional insights into Ubl domain-mediated regulation of SARS-CoV-2 PLpro",
"type": "journal-article",
"update-policy": "https://doi.org/10.1007/springer_crossmark_policy",
"volume": "20"
}