A Super-Resolution Spatial Atlas of SARS-CoV-2 Infection in Human Cells
et al., bioRxiv, doi:10.1101/2025.08.15.670620, Aug 2025
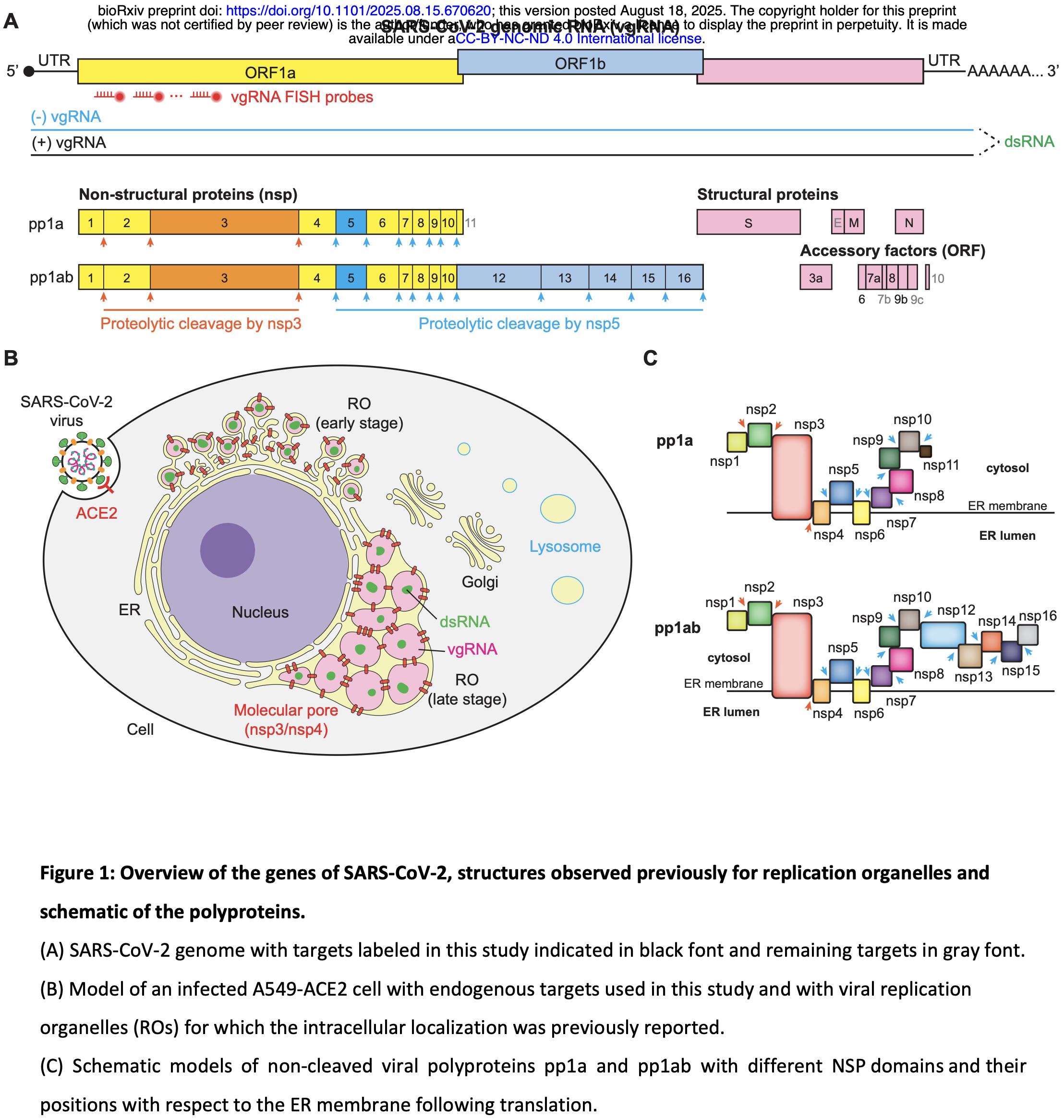
In vitro study showing the spatial organization of SARS-CoV-2 proteins and RNA in infected human lung cells using super-resolution microscopy. Authors used A549-ACE2 human lung epithelial cells infected with SARS-CoV-2 to create a nanoscale atlas mapping nearly all viral proteins and RNAs. The study reveals that the viral main protease nsp5 localizes inside double-membrane vesicles (DMVs) rather than in the cytoplasm as previously thought, suggesting polyprotein processing occurs after DMV formation.
Andronov et al., 18 Aug 2025, preprint, 6 authors.
Contact: wmoerner@stanford.edu, sqi@stanford.edu.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
A Super-Resolution Spatial Atlas of SARS-CoV-2 Infection in Human Cells
doi:10.1101/2025.08.15.670620
The spatial organization of viral and host components dictates the course of infection, yet the nanoscale architecture of the SARS-CoV-2 life cycle remains largely uncharted. Here, we present a comprehensive super-resolution Atlas of SARS-CoV-2 infection, systematically mapping the localization of nearly all viral proteins and RNAs in human cells. This resource reveals that the viral main protease, nsp5, localizes to the interior of double-membrane vesicles (DMVs), challenging existing models and suggesting that polyprotein processing is a terminal step in replication organelle maturation. We identify previously undescribed features of the infection landscape, including thin dsRNA "connectors" that physically link DMVs, and large, membrane-less dsRNA granules decorated with replicase components, reminiscent of viroplasms. Finally, we show that the antiviral drug nirmatrelvir induces the formation of persistent, multi-layered bodies of uncleaved polyproteins. This spatial Atlas provides a foundational resource for understanding coronavirus biology and offers crucial insights into viral replication, assembly, and antiviral mechanisms.
Confocal image analysis Image processing was performed in Fiji (ImageJ). Projections of adjacent Z planes showing maximum loci fluorescence were generated. For conditions 1 and 2 in Figure S4A , the number of infected cells and total cells were both manually counted using the "multi-point" function in Fiji. For condition 3, the number of infected cells was manually counted. Each field of view has several hundred cells in condition 3. To count the total number of cells, a Gaussian blur filter with sigma = 3.5 pixels was first applied to blur the fluorescence image of nsp4 and nsp5. The following functions in Fiji "Image->Adjust->Threshold", "Process->Binary->Fill Holes", "Analyze->Analyze particles" were sequentially performed to measure the cell number, with the size threshold set to greater than 30 µm 2 . The infection ratio was calculated using the number of infected cells divided by the total number of cells in each condition. Statistical analysis was performed using one-way ANOVA in GraphPad.
Optical setup for 2D 2-and 3-color SR microscopy 2D 2-and 3-color (d)STORM SR imaging was performed on a custom-built microscope 5 , using a Nikon Diaphot 200 inverted frame with an oil-immersion objective 60x/1.35 NA (Olympus UPLSAPO60XO) and a Si EMCCD camera (Andor iXon Ultra 897, 512×512 pixels). The sample was mounted on two stacked piezo stages (coarse U-780.DOS and fine P-545.3C8S, both Physik Instrumente). A 642 nm..
References
Alenquer, Vale-Costa, Etibor, Ferreira, Sousa et al., Influenza A virus ribonucleoproteins form liquid organelles at endoplasmic reticulum exit sites, Nature Communications, doi:10.1038/s41467-019-09549-4
Andronov, Genthial, Hentsch, Klaholz, splitSMLM, a spectral demixing method for high-precision multi-color localization microscopy applied to nuclear pore complexes, Communications Biology, doi:10.1038/s42003-022-04040-1
Andronov, Han, Zhu, Balaji, Roy et al., Nanoscale cellular organization of viral RNA and proteins in SARS-CoV-2 replication organelles, Nature Communications, doi:10.1038/s41467-024-48991-x
Andronov, Lutz, Vonesch, Klaholz, SharpViSu: integrated analysis and segmentation of super-resolution microscopy data, Bioinformatics, doi:10.1093/bioinformatics/btw123
Angeletti, Benvenuto, Bianchi, Giovanetti, Pascarella et al., COVID-2019: The role of the nsp2 and nsp3 in its pathogenesis, Journal of Medical Virology, doi:10.1002/jmv.25719
Angelini, Akhlaghpour, Neuman, Buchmeier, Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce doublemembrane vesicles, mBio, doi:10.1128/mBio.00524-13
Bou-Nader, Juma, Bothra, Brasington, Ghirlando et al., Specificity and mechanism of the double-stranded RNA-specific J2 monoclonal antibody, bioRxiv, doi:2009.649859.10.1101/2025.05.09.649859
Bouhamdani, Comeau, Turcotte, A Compendium of Information on the Lysosome, Front Cell Dev Biol, doi:10.3389/fcell.2021.798262
Braga, Barbosa De Souza, Pair-Pair Angular Correlation Function, doi:10.5772/67940
Chen, Lupan, Quek, Stanciu, Asaftei et al., A coronaviral porereplicase complex links RNA synthesis and export from double-membrane vesicles, Science Advances, doi:eadq9580.10.1126/sciadv.adq9580
Chiem, Ye, Martinez-Sobrido, Generation of Recombinant SARS-CoV-2 Using a Bacterial Artificial Chromosome, Current Protocols in Microbiology, doi:10.1002/cpmc.126
Chung, Irudayaraj, Lallow, Xu, Park et al., An overview of SARS-CoV-2 viral proteins with relevance to improved diagnostic and therapeutic platforms, Frontiers in Virology, doi:10.3389/fviro.2024.1399993
Cortese, Lee, Cerikan, Neufeldt, Oorschot et al., Integrative Imaging Reveals SARS-CoV-2-Induced Reshaping of Subcellular Morphologies, Cell Host & Microbe, doi:855.10.1016/j.chom.2020.11.003
Dahlberg, Saurabh, Sartor, Wang, Mitchell et al., Cryogenic single-molecule fluorescence annotations for electron tomography reveal in situ organization of key proteins in Caulobacter, Proceedings of the National Academy of Sciences of the United States of America, doi:10.1073/pnas.2001849117
Davies, Almasy, Mcdonald, Plate, Comparative Multiplexed Interactomics of SARS-CoV-2 and Homologous Coronavirus Nonstructural Proteins Identifies Unique and Shared Host-Cell Dependencies, ACS Infectious Diseases, doi:10.1021/acsinfecdis.0c00500
Doyle, Simpson, Hawes, Maier, Coronavirus RNA Synthesis Takes Place within Membrane-Bound Sites, Viruses
Ferreira, Fadl, Rabeh, Key dimer interface residues impact the catalytic activity of 3CLpro, the main protease of SARS-CoV-2, Journal of Biological Chemistry, doi:10.1016/j.jbc.2022.102023
Fisher, Gluck, Narayanan, Kuroda, Nachshon et al., Parsing the role of NSP1 in SARS-CoV-2 infection, Cell Reports, doi:10.1016/j.celrep.2022.110954
Gahlmann, Ptacin, Grover, Quirin, Von Diezmann et al., Quantitative Multicolor Subdiffraction Imaging of Bacterial Protein Ultrastructures in Three Dimensions, Nano Letters, doi:10.1021/nl304071h
Gao, Wang, Chen, Tepe, Huang et al., Perspectives on SARS-CoV-2 Main Protease Inhibitors, Journal of Medicinal Chemistry, doi:10.1021/acs.jmedchem.1c00409
Ghosh, Dellibovi-Ragheb, Kerviel, Pak, Qiu et al., β-Coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway, Cell
Gong, Qin, Dai, Tian, The glycosylation in SARS-CoV-2 and its receptor ACE2, Signal Transduction and Targeted Therapy, doi:10.1038/s41392-021-00809-8
Gordon, Jang, Bouhaddou, Xu, Obernier et al., A SARS-CoV-2 protein interaction map reveals targets for drug repurposing, Nature, doi:10.1038/s41586-020-2286-9
Grover, Pavani, Piestun, Performance limits on three-dimensional particle localization in photon-limited microscopy, Opt. Lett
Guseva, Milles, Jensen, Salvi, Kleman et al., Measles virus nucleo-and phosphoproteins form liquid-like phaseseparated compartments that promote nucleocapsid assembly, Science Advances, doi:10.1126/sciadv.aaz7095
Hagemeijer Marne, Verheije Monique, Ulasli, Shaltiël Indra, De Vries Lisa et al., Dynamics of Coronavirus Replication-Transcription Complexes, Journal of Virology, doi:10.1128/jvi.01716-09
Han, Zhou, Liu, Wang, Qin et al., SARS-CoV-2 N protein coordinates viral particle assembly through multiple domains, Journal of Virology, doi:10.1128/jvi.01036-24
Hartenian, Nandakumar, Lari, Ly, Tucker et al., The molecular virology of coronaviruses, J Biol Chem, doi:10.1074/jbc.REV120.013930
Helmerich, Beliu, Matikonda, Schnermann, Sauer, Photoblueing of organic dyes can cause artifacts in super-resolution microscopy, Nature Methods, doi:10.1038/s41592-021-01061-2
Hoffmann, Kleine-Weber, Pöhlmann, A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells, Molecular Cell, doi:775.10.1016/j.molcel.2020.04.022
Huang, Wang, Zhong, Zhang, Zhang et al., Molecular architecture of coronavirus double-membrane vesicle pore complex, Nature, doi:10.1038/s41586-024-07817-y
Is A Sarafan Chem-H Fellow, Liu, Liu, Teng, Wang et al., acknowledge support by the Stanford School of Medicine Dean's Postdoctoral Fellowship, Signal Transduction and Targeted Therapy, doi:10.1038/s41392-020-00372-8
Islam, Islam, Siddik Alom, Kabir, Halim, A review on structural, non-structural, and accessory proteins of SARS-CoV-2: Highlighting drug target sites, Immunobiology, doi:10.1016/j.imbio.2022.152302
Izadpanah, Rappaport, Datta, Epitranscriptomics of SARS-CoV-2 Infection, Frontiers in Cell and Developmental Biology, doi:10.3389/fcell.2022.849298
Kai, Kai, Interactions of coronaviruses with ACE2, angiotensin II, and RAS inhibitors-lessons from available evidence and insights into COVID-19, Hypertension Research, doi:10.1038/s41440-020-0455-8
Kim, Park, Yoon, Lee, Kim et al., SARS-CoV-2 Nsp2 recruits GIGYF2 near viral replication sites and supports viral protein production, Nucleic Acids Research, doi:10.1093/nar/gkaf674
Klein, Cortese, Winter, Wachsmuth-Melm, Neufeldt et al., SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography, Nat Commun, doi:10.1038/s41467-020-19619-7
Knoops, Kikkert, Worm, Zevenhoven-Dobbe, Van Der Meer et al., SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum, PLoS Biol, doi:10.1371/journal.pbio.0060226
Lee, Wing, Gala, Noerenberg, Järvelin et al., Absolute quantitation of individual SARS-CoV-2 RNA molecules provides a new paradigm for infection dynamics and variant differences, eLife, doi:10.7554/eLife.74153
Liv, Xiaohan, Jana, Moritz, Vibhu et al., SARS-CoV-2 nsp3-4 suffice to form a pore shaping replication organelles, bioRxiv, doi:2010.2021.513196.10.1101/2022.10.21.513196
Malone, Urakova, Snijder, Campbell, Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design, Nature Reviews Molecular Cell Biology, doi:10.1038/s41580-021-00432-z
Marano, Vlachová, Tiano, A portrait of the infected cell: how SARS-CoV-2 infection reshapes cellular processes and pathways, npj Viruses, doi:10.1038/s44298-024-00076-8
Martens, Bader, Baas, Rieger, Hohlbein, Phasor based single-molecule localization microscopy in 3D (pSMLM-3D): An algorithm for MHz localization rates using standard CPUs, The Journal of chemical physics, doi:10.1063/1.5005899;2410.1063/1.5005899
Mei, Cupic, Miorin, Ye, Cagatay et al., Inhibition of mRNA nuclear export promotes SARS-CoV-2 pathogenesis, Proceedings of the National Academy of Sciences, doi:10.1073/pnas.2314166121
Mendonça, Howe, Gilchrist, Sheng, Sun et al., Correlative multi-scale cryo-imaging unveils SARS-CoV-2 assembly and egress, Nature Communications, doi:10.1038/s41467-021-24887-y
Mukherjee, Dikic, Proteases of SARS Coronaviruses, doi:10.1016/B978-0-12-821618-7.00111-5
Möckl, Super-resolution microscopy with single molecules in biology and beyond -essentials, current trends, and future challenges, J. Am. Chem. Soc
Oudshoorn, Rijs, Limpens Ronald, Groen, Koster Abraham et al., Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication, mBio, doi:10.1128/mbio.01658-01617.10.1128/mbio.01658-17
Ovesny, Krizek, Borkovec, Svindrych, Hagen, ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging, Bioinformatics, doi:10.1093/bioinformatics/btu202
Owen, Allerton, Anderson, Aschenbrenner, Avery et al., An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19, Science, doi:10.1126/science.abl4784
Papa, Borodavka, Desselberger, Viroplasms: Assembly and Functions of Rotavirus Replication Factories, Viruses
Pavani, Thompson, Biteen, Lord, Liu et al., Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function, Proceedings of the National Academy of Sciences of the United States of America, doi:10.1073/pnas.0900245106
Rachel, Sims Amy, Brockway Sarah, Baric Ralph, Mark, The nsp2 Replicase Proteins of Murine Hepatitis Virus and Severe Acute Respiratory Syndrome Coronavirus Are Dispensable for Viral Replication, Journal of Virology, doi:10.1128/jvi.79.21.13399-13411.2005
Razvag, Neve-Oz, Sajman, Reches, Sherman, Nanoscale kinetic segregation of TCR and CD45 in engaged microvilli facilitates early T cell activation, Nature Communications, doi:10.1038/s41467-018-03127-w
Ricciardi, Guarino, Giaquinto, Polishchuk, Santoro et al., The role of NSP6 in the biogenesis of the SARS-CoV-2 replication organelle, Nature, doi:10.1038/s41586-022-04835-6
Ries, Kaplan, Platonova, Eghlidi, Ewers, A simple, versatile method for GFP-based super-resolution microscopy via nanobodies, Nat. Methods, doi:10.1038/NMETH.1991
Rihn, Merits, Bakshi, Turnbull, Wickenhagen et al., A plasmid DNA-launched SARS-CoV-2 reverse genetics system and coronavirus toolkit for COVID-19 research, PLOS Biology, doi:10.1371/journal.pbio.3001091
Rust, Bates, Zhuang, Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM), Nat. Methods
Scherer, Mascheroni, Carnell, Wunderlich, Makarchuk et al., SARS-CoV-2 nucleocapsid protein adheres to replication organelles before viral assembly at the Golgi/ERGIC and lysosome-mediated egress, Science Advances, doi:10.1126/sciadv.abl4895
Schindelin, Arganda-Carreras, Frise, Kaynig, Longair et al., Fiji: an open-source platform for biological-image analysis, Nature methods
Schonborn, Oberstrass, Breyel, Tittgen, Schumacher et al., Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts, Nucleic Acids Res, doi:10.1093/nar/19.11.2993
Schubert, Karousis, Jomaa, Scaiola, Echeverria et al., SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation, Nature Structural & Molecular Biology, doi:10.1038/s41594-020-0511-8
Schueder, Rivera-Molina, Su, Marin, Kidd et al., Unraveling cellular complexity with transient adapters in highly multiplexed super-resolution imaging, Cell, doi:1718.10.1016/j.cell.2024.02.033
Shi, Yu, Li, Cui, Zhao et al., Expression Profile and Localization of SARS-CoV-2 Nonstructural Replicase Proteins in Infected Cells, Microbiology Spectrum, doi:10.1128/spectrum.00744-22
Snijder, Limpens, De Wilde, Jong, Zevenhoven-Dobbe et al., A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis, PLoS Biol, doi:10.1371/journal.pbio.3000715
Steiner, Kratzel, Barut, Lang, Aguiar Moreira et al., SARS-CoV-2 biology and host interactions, Nature Reviews Microbiology, doi:10.1038/s41579-023-01003-z
Stertz, Reichelt, Spiegel, Kuri, Martinez-Sobrido et al., The intracellular sites of early replication and budding of SARScoronavirus, Virology, doi:10.1016/j.virol.2006.11.027
Sundquist, Kräusslich, HIV-1 Assembly, Budding, and Maturation, Cold Spring Harbor Perspectives in Medicine, doi:10.1101/cshperspect.a006924
Van De Linde, Loeschberger, Klein, Heidbreder, Wolter et al., Direct stochastic optical reconstruction microscopy with standard fluorescent probes, Nature Protocols
Van Hemert, Van Den Worm, Knoops, Mommaas, Gorbalenya et al., SARS-Coronavirus Replication/Transcription Complexes Are Membrane-Protected and Need a Host Factor for Activity In Vitro, PLOS Pathogens, doi:10.1371/journal.ppat.1000054
Von Diezmann, Shechtman, Moerner, Three-Dimensional Localization of Single Molecules for Super-Resolution Imaging and Single-Particle Tracking, Chemical Reviews, doi:10.1021/acs.chemrev.6b00629
Wang, Xiong, Susanto, Li, Leung et al., Transforming Rhodamine Dyes for (d)STORM Super-Resolution Microscopy via 1,3-Disubstituted Imidazolium Substitution, Angewandte Chemie International Edition, doi:10.1002/anie.202113612
Westberg, Su, Zou, Huang, Rustagi et al., An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations, Science Translational Medicine, doi:10.1126/scitranslmed.adi0979
Wolff, Limpens, Zevenhoven-Dobbe, Laugks, Zheng et al., A molecular pore spans the double membrane of the coronavirus replication organelle, Science, doi:10.1126/science.abd3629
Wu, Xiao, Liu, Wen, Jin et al., Dynamics of RNA localization to nuclear speckles are connected to splicing efficiency, Science Advances, doi:10.1126/sciadv.adp7727
Yang, Tian, Wang, Chen, Xiao et al., SARS-CoV-2 NSP3/4 control formation of replication organelle and recruitment of RNA polymerase NSP12, Journal of Cell Biology, doi:10.1083/jcb.202306101
Yao, Song, Chen, Wu, Xu et al., Molecular Architecture of the SARS-CoV-2 Virus, Cell, doi:10.1016/j.cell.2020.09.018
Ye, Chiem, Park, Oladunni, Platt Roy et al., Rescue of SARS-CoV-2 from a Single Bacterial Artificial Chromosome, mBio, doi:10.1128/mbio.02168-02120.10.1128/mbio.02168-20
Zhang, Kennedy, De Melo Jorge, Xing, Reid et al., SARS-CoV-2 remodels the Golgi apparatus to facilitate viral assembly and secretion, PLOS Pathogens, doi:10.1371/journal.ppat.1013295
Zhang, Miorin, Makio, Dehghan, Gao et al., Nsp1 protein of SARS-CoV-2 disrupts the mRNA export machinery to inhibit host gene expression, Science Advances, doi:10.1126/sciadv.abe7386
Zhang, Xu, Bai, Chen, Cates et al., A subcellular map of translational machinery composition and regulation at the single-molecule level, Science, doi:2623.10.1126/science.adn2623
Zhu, Balaji, Han, Andronov, Roy et al., High-resolution dynamic imaging of chromatin DNA communication using Oligo-LiveFISH, Cell, doi:10.1016/j.cell.2025.03.032
Zimmermann, Zhao, Makroczyova, Wachsmuth-Melm, Prasad et al., SARS-CoV-2 nsp3 and nsp4 are minimal constituents of a pore spanning replication organelle, Nature Communications, doi:10.1038/s41467-023-43666-5
DOI record:
{
"DOI": "10.1101/2025.08.15.670620",
"URL": "http://dx.doi.org/10.1101/2025.08.15.670620",
"abstract": "<jats:title>Abstract</jats:title>\n <jats:p>The spatial organization of viral and host components dictates the course of infection, yet the nanoscale architecture of the SARS-CoV-2 life cycle remains largely uncharted. Here, we present a comprehensive super-resolution Atlas of SARS-CoV-2 infection, systematically mapping the localization of nearly all viral proteins and RNAs in human cells. This resource reveals that the viral main protease, nsp5, localizes to the interior of double-membrane vesicles (DMVs), challenging existing models and suggesting that polyprotein processing is a terminal step in replication organelle maturation. We identify previously undescribed features of the infection landscape, including thin dsRNA “connectors” that physically link DMVs, and large, membrane-less dsRNA granules decorated with replicase components, reminiscent of viroplasms. Finally, we show that the antiviral drug nirmatrelvir induces the formation of persistent, multi-layered bodies of uncleaved polyproteins. This spatial Atlas provides a foundational resource for understanding coronavirus biology and offers crucial insights into viral replication, assembly, and antiviral mechanisms.</jats:p>",
"accepted": {
"date-parts": [
[
2025,
8,
18
]
]
},
"author": [
{
"ORCID": "https://orcid.org/0000-0002-4213-5363",
"affiliation": [],
"authenticated-orcid": false,
"family": "Andronov",
"given": "Leonid",
"sequence": "first"
},
{
"affiliation": [],
"family": "Han",
"given": "Mengting",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Balaji",
"given": "Ashwin",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zhu",
"given": "Yanyu",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-3965-3223",
"affiliation": [],
"authenticated-orcid": false,
"family": "Qi",
"given": "Lei S.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-2830-209X",
"affiliation": [],
"authenticated-orcid": false,
"family": "Moerner",
"given": "W. E.",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
8,
19
]
],
"date-time": "2025-08-19T03:55:19Z",
"timestamp": 1755575719000
},
"deposited": {
"date-parts": [
[
2025,
8,
21
]
],
"date-time": "2025-08-21T14:25:17Z",
"timestamp": 1755786317000
},
"funder": [
{
"DOI": "10.13039/100000057",
"award": [
"R35GM118067"
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/100000057",
"id-type": "DOI"
}
],
"name": "National Institute of General Medical Sciences"
},
{
"award": [
"U01 DK127405"
],
"name": "National Institutes of Health Common Fund 4D Nucleome Program"
}
],
"group-title": "Microbiology",
"indexed": {
"date-parts": [
[
2025,
8,
22
]
],
"date-time": "2025-08-22T02:10:44Z",
"timestamp": 1755828644978,
"version": "3.44.0"
},
"institution": [
{
"name": "bioRxiv"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2025,
8,
18
]
]
},
"license": [
{
"URL": "http://creativecommons.org/licenses/by-nc-nd/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
8,
18
]
],
"date-time": "2025-08-18T00:00:00Z",
"timestamp": 1755475200000
}
}
],
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2025.08.15.670620",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2025,
8,
18
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2025,
8,
18
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference": [
{
"DOI": "10.1038/s41392-020-00372-8",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.1"
},
{
"DOI": "10.1128/spectrum.00744-22",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.2"
},
{
"DOI": "10.1371/journal.pbio.3001091",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.3"
},
{
"DOI": "10.1038/s41586-024-07817-y",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.4"
},
{
"DOI": "10.1038/s41467-024-48991-x",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.5"
},
{
"DOI": "10.3390/v13122540",
"article-title": "Coronavirus RNA Synthesis Takes Place within Membrane-Bound Sites",
"doi-asserted-by": "crossref",
"first-page": "2540",
"journal-title": "Viruses",
"key": "2025082107250834000_2025.08.15.670620v1.6",
"volume": "13",
"year": "2021"
},
{
"DOI": "10.1371/journal.pbio.3000715",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.7"
},
{
"DOI": "10.1126/sciadv.adq9580",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.8"
},
{
"DOI": "10.1083/jcb.202306101",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.9"
},
{
"DOI": "10.1021/jacs.0c08178",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.10"
},
{
"DOI": "10.1038/nmeth929",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.11"
},
{
"DOI": "10.1016/j.chom.2020.11.003",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.12"
},
{
"DOI": "10.1038/s41579-023-01003-z",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.13"
},
{
"DOI": "10.1038/s41440-020-0455-8",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.14"
},
{
"DOI": "10.1038/nprot.2011.336",
"article-title": "Direct stochastic optical reconstruction microscopy with standard fluorescent probes",
"doi-asserted-by": "crossref",
"first-page": "991",
"journal-title": "Nature Protocols",
"key": "2025082107250834000_2025.08.15.670620v1.15",
"volume": "6",
"year": "2011"
},
{
"DOI": "10.1021/acs.chemrev.6b00629",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.16"
},
{
"DOI": "10.1038/s42003-022-04040-1",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.17"
},
{
"DOI": "10.1002/anie.202113612",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.18"
},
{
"DOI": "10.1093/nar/19.11.2993",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.19"
},
{
"DOI": "10.1101/2025.05.09.649859",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.20"
},
{
"DOI": "10.7554/eLife.74153",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.21"
},
{
"DOI": "10.1016/B978-0-12-821618-7.00111-5",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.22"
},
{
"DOI": "10.1038/s41594-020-0511-8",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.23"
},
{
"DOI": "10.1126/sciadv.abe7386",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.24"
},
{
"DOI": "10.1073/pnas.2314166121",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.25"
},
{
"DOI": "10.1016/j.celrep.2022.110954",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.26"
},
{
"DOI": "10.1126/science.adn2623",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.27"
},
{
"DOI": "10.1021/acsinfecdis.0c00500",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.28"
},
{
"DOI": "10.1038/s41586-020-2286-9",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.29"
},
{
"DOI": "10.1093/nar/gkaf674",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.30"
},
{
"DOI": "10.1128/JVI.79.21.13399-13411.2005",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.31"
},
{
"DOI": "10.1371/journal.pbio.0060226",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.32"
},
{
"DOI": "10.1128/JVI.01716-09",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.33"
},
{
"DOI": "10.1371/journal.ppat.1013295",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.34"
},
{
"DOI": "10.1021/acs.jmedchem.1c00409",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.35"
},
{
"DOI": "10.1126/science.abl4784",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.36"
},
{
"DOI": "10.1074/jbc.REV120.013930",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.37"
},
{
"DOI": "10.1038/s41580-021-00432-z",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.38"
},
{
"DOI": "10.1038/s44298-024-00076-8",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.39"
},
{
"DOI": "10.1038/s41467-023-43666-5",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.40"
},
{
"DOI": "10.1101/2022.10.21.513196",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.41"
},
{
"DOI": "10.1126/science.abd3629",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.42"
},
{
"DOI": "10.1038/s41467-020-19619-7",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.43"
},
{
"DOI": "10.1128/mBio.00524-13",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.44"
},
{
"DOI": "10.1038/s41586-022-04835-6",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.45"
},
{
"DOI": "10.1371/journal.ppat.1000054",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.46"
},
{
"DOI": "10.5772/67940",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.47"
},
{
"DOI": "10.1073/pnas.0900245106",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.48"
},
{
"DOI": "10.1364/OL.35.003306",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.49"
},
{
"DOI": "10.1038/nmeth.1991",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.50"
},
{
"DOI": "10.1016/j.imbio.2022.152302",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.51"
},
{
"DOI": "10.3389/fviro.2024.1399993",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.52"
},
{
"DOI": "10.1016/j.cell.2020.09.018",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.53"
},
{
"DOI": "10.1038/s41392-021-00809-8",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.54"
},
{
"DOI": "10.1016/j.cell.2024.02.033",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.55"
},
{
"DOI": "10.1016/J.CELL.2020.10.039",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.56"
},
{
"DOI": "10.3389/fcell.2021.798262",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.57"
},
{
"DOI": "10.1126/sciadv.abl4895",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.58"
},
{
"DOI": "10.1016/j.molcel.2020.04.022",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.59"
},
{
"DOI": "10.1016/j.jbc.2022.102023",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.60"
},
{
"DOI": "10.1101/cshperspect.a006924",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.61"
},
{
"DOI": "10.1002/jmv.25719",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.62"
},
{
"DOI": "10.3390/v13071349",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.63"
},
{
"DOI": "10.1126/sciadv.aaz7095",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.64"
},
{
"DOI": "10.1038/s41467-019-09549-4",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.65"
},
{
"DOI": "10.1126/sciadv.adp7727",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.66"
},
{
"DOI": "10.1016/j.virol.2006.11.027",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.67"
},
{
"DOI": "10.1038/s41467-021-24887-y",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.68"
},
{
"DOI": "10.1128/jvi.01036-24",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.69"
},
{
"DOI": "10.1128/mbio.01658-01617",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.70"
},
{
"DOI": "10.3389/fcell.2022.849298",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.71"
},
{
"DOI": "10.1073/pnas.2001849117",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.72"
},
{
"DOI": "10.1002/cpmc.126",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.73"
},
{
"DOI": "10.1128/mbio.02168-02120",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.74"
},
{
"DOI": "10.1126/scitranslmed.adi0979",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.75"
},
{
"DOI": "10.1093/bioinformatics/btu202",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.76"
},
{
"DOI": "10.1063/1.5005899",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.77"
},
{
"DOI": "10.1038/nmeth.2019",
"article-title": "Fiji: an open-source platform for biological-image analysis",
"doi-asserted-by": "crossref",
"first-page": "676",
"journal-title": "Nature methods",
"key": "2025082107250834000_2025.08.15.670620v1.78",
"volume": "9",
"year": "2012"
},
{
"DOI": "10.1093/bioinformatics/btw123",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.79"
},
{
"DOI": "10.1038/s41592-021-01061-2",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.80"
},
{
"DOI": "10.1016/j.cell.2025.03.032",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.81"
},
{
"DOI": "10.1021/nl304071h",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.82"
},
{
"DOI": "10.1038/s41467-018-03127-w",
"doi-asserted-by": "publisher",
"key": "2025082107250834000_2025.08.15.670620v1.83"
}
],
"reference-count": 83,
"references-count": 83,
"relation": {},
"resource": {
"primary": {
"URL": "http://biorxiv.org/lookup/doi/10.1101/2025.08.15.670620"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"subtype": "preprint",
"title": "A Super-Resolution Spatial Atlas of SARS-CoV-2 Infection in Human Cells",
"type": "posted-content"
}