TransFactor—prediction of pro-viral SARS-CoV-2 host factors using a protein language model
et al., Bioinformatics, doi:10.1093/bioinformatics/btaf491, Sep 2025
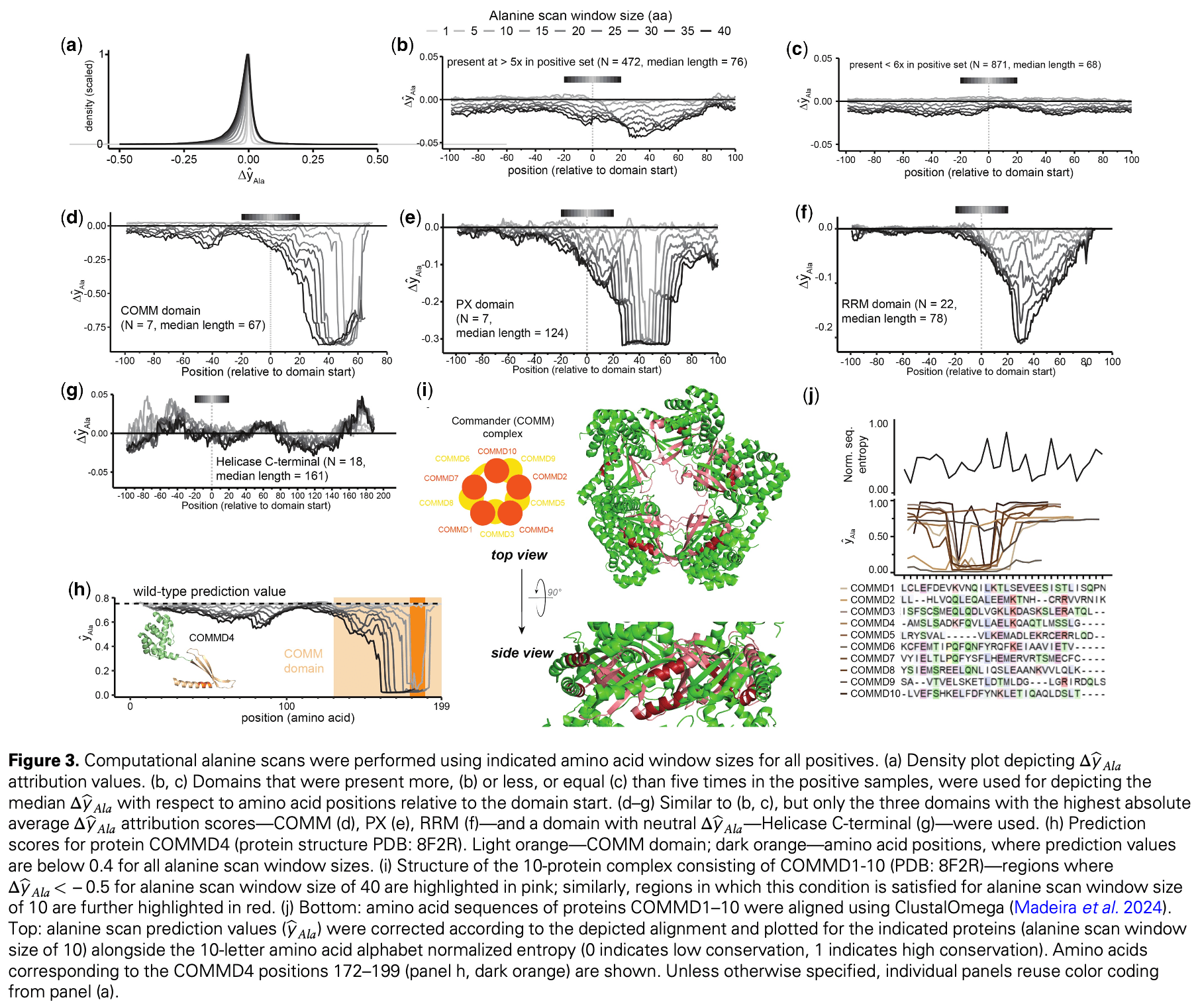
In silico study presenting a computational framework to predict and prioritize pro-viral SARS-CoV-2 host factors using only protein sequence data. Through a computational alanine scan used for model interpretation, authors identified specific pro-viral protein domains as potential therapeutic targets, including copper metabolism gene MURR1 (COMM), phox homology (PX), and RNA recognition motifs (RRM).
An et al., 1 Sep 2025, peer-reviewed, 9 authors.
Contact: annalisa.marsico@helmholtz-munich.de.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Sequence analysis TransFactor-prediction of pro-viral SARS-CoV-2 host factors using a protein language model
Motivation: Recent pandemics have revealed significant gaps in our understanding of viral pathogenesis, exposing an urgent need for methods to identify and prioritize key host proteins (host factors) as potential targets for antiviral treatments. De novo generation of experimental datasets is limited by their heterogeneity, and for looming future pandemics, may not be feasible due to limitations of experimental approaches. Results: Here, we present TransFactor, a computational framework for predicting and prioritizing candidate host factors using only protein sequence data. It leverages the pre-trained ESM-2 protein language model, fine-tuned on a limited set of experimentally determined host factors aggregated from 33 independent SARS-CoV-2 studies. TransFactor outperforms machine and deep learning baselines and its predictions align with Gene Ontology enrichments of known host factors, but also provide interpretability through a computational alanine scan, enabling the identification of pro-viral protein domains such as COMM, PX, and RRM, that may be used to direct experimental investigations of virus biology and guide rational design of antiviral therapies. Our findings demonstrate the potential of transformer-based models to advance host factor prediction, providing a framework extendable to orthogonal input modalities and other infectious diseases, enhancing our preparedness for current and future viral threats.
Author contributions Yang An (Conceptualization [equal]
References
Akiba, Sano, Yanase, Optuna: a next-generation hyperparameter optimization framework
Baggen, Vanstreels, Jansen, Cellular host factors for SARS-CoV-2 infection, Nat Microbiol
Bergant, Yamada, Grass, Attenuation of SARS-CoV-2 replication and associated inflammation by concomitant targeting of viral and host cap 2 0 -o-ribose methyltransferases, EMBO J
Bressin, Schulte-Sasse, Figini, TriPepSVM: de novo prediction of RNA-binding proteins based on short amino acid motifs, Nucleic Acids Res
Dao, Fu, Ermon, FlashAttention: fast and memory-efficient exact attention with io-awareness, Adv Neural Inf Process Syst
De Clercq, Li, Approved antiviral drugs over the past 50 years, Clin Microbiol Rev
Devlin, Chang, Lee, Pre-training of deep bidirectional transformers for language understanding
Dhodapkar, A deep generative model of the SARS-CoV-2 spike protein predicts future variants, bioRxiv
Ferrarini, Lal, Rebollo, Genome-wide bioinformatic analyses predict key host and viral factors in SARS-CoV-2 pathogenesis, Commun Biol
Gysi, Valle � I, Zitnik, Network medicine framework for identifying drug-repurposing opportunities for covid-19, Proc Natl Acad Sci
Healy, Mcnally, Butkovi� C R, Structure of the endosomal commander complex linked to Ritscher-Schinzel syndrome, Cell
Hie, Yang, Kim, Evolutionary velocity with protein language models predicts evolutionary dynamics of diverse proteins, Cell Syst
Horlacher, Oleshko, Hu, A computational map of the human-SARS-CoV-2 protein-RNA interactome predicted at singlenucleotide resolution, NAR Genom Bioinform
Hu, Shen, Wallis, Low-rank adaptation of large language models, ICLR
Huang, Bergant, Grass, Multi-omics characterization of the monkeypox virus infection, Nat Commun
Kaufmann, Dorhoi, Hotchkiss, Host-directed therapies for bacterial and viral infections, Nat Rev Drug Discov
Kortemme, Kim, Baker, Computational alanine scanning of protein-protein interfaces, Sci STKE
Li, Li, Li, Machine learning early detection of SARS-CoV-2 high-risk variants, Adv Sci
Lin, Akin, Rao, Evolutionary-scale prediction of atomiclevel protein structure with a language model, Science
Liu, Nie, Si, Generative prediction of real-world prevalent SARS-CoV-2 mutation with in silico virus evolution, Brief Bioinform
Madeira, Madhusoodanan, Lee, The EMBL-EBI job dispatcher sequence analysis tools framework in 2024, Nucleic Acids Res
Massova, Kollman, Computational alanine scanning to probe protein-protein interactions: a novel approach to evaluate binding free energies, J Am Chem Soc
Montoya, Ready, A virus-packageable CRISPR system identifies host dependency factors co-opted by multiple HIV-1 strains, mBio
Morrison, Weiss, Combinatorial alanine-scanning, Curr Opin Chem Biol
Mosharaf, Reza, Kibria, Computational identification of host genomic biomarkers highlighting their functions, pathways and regulators that influence SARS-CoV-2 infections and drug repurposing, Sci Rep
Rancati, Nicora, Bergomi, SARITA: a large language model for generating the S1 subunit of the SARS-CoV-2 spike protein, Brief Bioinform
Rancati, Nicora, Prosperi, Forecasting dominance of SARS-CoV-2 lineages by anomaly detection using deep autoencoders, Brief Bioinform
Ravindran, Wagoner, Athanasiadis, Discovery of hostdirected modulators of virus infection by probing the SARS-CoV-2host protein-protein interaction network, Brief Bioinform
Rebendenne, Roy, Bonaventure, Bidirectional genome-wide CRISPR screens reveal host factors regulating SARS-CoV-2, MERS-CoV and seasonal HCoVs, Nat Genet
Ruiz, Zitnik, Leskovec, Identification of disease treatment mechanisms through the multiscale interactome, Nat Commun
Samy, Maher, Abdelsalam, SARS-CoV-2 potential drugs, drug targets, and biomarkers: a viral-host interaction network-based analysis, Sci Rep
Scaturro, Stukalov, Haas, An orthogonal proteomic survey uncovers novel Zika virus host factors, Nature
Schmirler, Heinzinger, Rost, Fine-tuning protein language models boosts predictions across diverse tasks, Nat Commun
Sherman, Hao, Qiu, DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update), Nucleic Acids Res
Steinegger, MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets, Nat Biotechnol
Stukalov, Girault, Grass, Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV, Nature
Szklarczyk, Kirsch, Koutrouli, The string database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest, Nucleic Acids Res
Thadani, Gurev, Notin, Learning from prepandemic data to forecast viral escape, Nature
Tiwari, Mishra, Gupta, Structural similarity-based prediction of host factors associated with SARS-CoV-2 infection and pathogenesis, J Biomol Struct Dyn
Vandelli, Monti, Milanetti, Structural analysis of SARS-CoV-2 genome and predictions of the human interactome, Nucleic Acids Res
Vaswani, Shazeer, Parmar, Attention is all you need, NIPS
Wang, Li, Zhang, A comprehensive review of protein language models
Wu, Guo, Improved prediction of DNA and RNA binding proteins with deep learning models, Brief Bioinform
Xiao, Zhao, Zhang, Protein large language models: a comprehensive survey
Youssef, Gurev, Ghantous, Computationally designed proteins mimic antibody immune evasion in viral evolution, Immunity
Zhu, Feng, Hu, A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry, Nat Commun
DOI record:
{
"DOI": "10.1093/bioinformatics/btaf491",
"ISSN": [
"1367-4811"
],
"URL": "http://dx.doi.org/10.1093/bioinformatics/btaf491",
"abstract": "<jats:title>Abstract</jats:title>\n <jats:sec>\n <jats:title>Motivation</jats:title>\n <jats:p>Recent pandemics have revealed significant gaps in our understanding of viral pathogenesis, exposing an urgent need for methods to identify and prioritize key host proteins (host factors) as potential targets for antiviral treatments. De novo generation of experimental datasets is limited by their heterogeneity, and for looming future pandemics, may not be feasible due to limitations of experimental approaches.</jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>Results</jats:title>\n <jats:p>Here, we present TransFactor, a computational framework for predicting and prioritizing candidate host factors using only protein sequence data. It leverages the pre-trained ESM-2 protein language model, fine-tuned on a limited set of experimentally determined host factors aggregated from 33 independent SARS-CoV-2 studies. TransFactor outperforms machine and deep learning baselines and its predictions align with Gene Ontology enrichments of known host factors, but also provide interpretability through a computational alanine scan, enabling the identification of pro-viral protein domains such as COMM, PX, and RRM, that may be used to direct experimental investigations of virus biology and guide rational design of antiviral therapies. Our findings demonstrate the potential of transformer-based models to advance host factor prediction, providing a framework extendable to orthogonal input modalities and other infectious diseases, enhancing our preparedness for current and future viral threats.</jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>Availability and implementation</jats:title>\n <jats:p>Source code is available at https://github.com/marsico-lab/TransFactor. A full reproducibility package, including code, trained models, and data, is archived on Zenodo (https://doi.org/10.5281/zenodo.16793684).</jats:p>\n </jats:sec>",
"article-number": "btaf491",
"author": [
{
"ORCID": "https://orcid.org/0000-0003-1225-3534",
"affiliation": [
{
"name": "Computational Health Center, Helmholtz Center Munich , Neuherberg 85764,",
"place": [
"Germany"
]
},
{
"name": "School of Computation, Information and Technology, Technical University of Munich , Munich 80333,",
"place": [
"Germany"
]
}
],
"authenticated-orcid": false,
"family": "An",
"given": "Yang",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0000-0003-3458-9506",
"affiliation": [
{
"name": "Institute of Virology, Technical University of Munich , Munich 80333,",
"place": [
"Germany"
]
},
{
"name": "Department of Molecular Biology and Nanobiotechnology, National Institute of Chemistry , Ljubljana 1000,",
"place": [
"Slovenia"
]
}
],
"authenticated-orcid": false,
"family": "Bergant",
"given": "Valter",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Computational Health Center, Helmholtz Center Munich , Neuherberg 85764,",
"place": [
"Germany"
]
},
{
"name": "School of Computation, Information and Technology, Technical University of Munich , Munich 80333,",
"place": [
"Germany"
]
}
],
"family": "Firmani",
"given": "Samuele",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Virology, Technical University of Munich , Munich 80333,",
"place": [
"Germany"
]
}
],
"family": "Grünke",
"given": "Corinna",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Virology, Technical University of Munich , Munich 80333,",
"place": [
"Germany"
]
}
],
"family": "Bonnal",
"given": "Batiste",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Virology, Technical University of Munich , Munich 80333,",
"place": [
"Germany"
]
}
],
"family": "Henrici",
"given": "Alexander",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-0166-1367",
"affiliation": [
{
"name": "Institute of Virology, Technical University of Munich , Munich 80333,",
"place": [
"Germany"
]
},
{
"name": "German Center for Infection Research (DZIF), Munich Partner Site , Munich 81675,",
"place": [
"Germany"
]
},
{
"name": "Systems Virology, Helmholtz Center Munich , Neuherberg 85764,",
"place": [
"Germany"
]
}
],
"authenticated-orcid": false,
"family": "Pichlmair",
"given": "Andreas",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-3412-1102",
"affiliation": [
{
"name": "Computational Health Center, Helmholtz Center Munich , Neuherberg 85764,",
"place": [
"Germany"
]
}
],
"authenticated-orcid": false,
"family": "Schubert",
"given": "Benjamin",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Computational Health Center, Helmholtz Center Munich , Neuherberg 85764,",
"place": [
"Germany"
]
}
],
"family": "Marsico",
"given": "Annalisa",
"sequence": "additional"
}
],
"container-title": "Bioinformatics",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
9,
9
]
],
"date-time": "2025-09-09T11:31:54Z",
"timestamp": 1757417514000
},
"deposited": {
"date-parts": [
[
2025,
9,
20
]
],
"date-time": "2025-09-20T01:10:25Z",
"timestamp": 1758330625000
},
"editor": [
{
"affiliation": [],
"family": "Cheng",
"given": "Jianlin",
"sequence": "additional"
}
],
"funder": [
{
"name": "BMBF Clusters4Future"
},
{
"name": "CNATM"
},
{
"name": "Helmholtz International Lab Causal Cell Dynamics"
},
{
"DOI": "10.13039/501100001656",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100001656",
"id-type": "DOI"
}
],
"name": "Helmholtz Association"
},
{
"award": [
"TRR179/TP10",
"TRR237/A07",
"TRR353/B04"
],
"name": "the German Research Foundation"
},
{
"award": [
"KA1-Co-02"
],
"name": "Helmholtz Association’s Initiative and Networking Fund"
},
{
"name": "Center for Immunology of Viral Infections"
},
{
"DOI": "10.13039/501100001732",
"award": [
"164"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100001732",
"id-type": "DOI"
}
],
"name": "Danish National Research Foundation"
},
{
"DOI": "10.13039/501100004329",
"award": [
"P1-0242",
"J4-60082"
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100004329",
"id-type": "DOI"
}
],
"name": "Slovenian Research and Innovation Agency"
}
],
"indexed": {
"date-parts": [
[
2025,
9,
20
]
],
"date-time": "2025-09-20T06:29:31Z",
"timestamp": 1758349771149,
"version": "3.44.0"
},
"is-referenced-by-count": 0,
"issue": "9",
"issued": {
"date-parts": [
[
2025,
9,
1
]
]
},
"journal-issue": {
"issue": "9",
"published-print": {
"date-parts": [
[
2025,
9,
1
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 9,
"start": {
"date-parts": [
[
2025,
9,
10
]
],
"date-time": "2025-09-10T00:00:00Z",
"timestamp": 1757462400000
}
}
],
"link": [
{
"URL": "https://academic.oup.com/bioinformatics/advance-article-pdf/doi/10.1093/bioinformatics/btaf491/64237229/btaf491.pdf",
"content-type": "application/pdf",
"content-version": "am",
"intended-application": "syndication"
},
{
"URL": "https://academic.oup.com/bioinformatics/article-pdf/41/9/btaf491/64237229/btaf491.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "syndication"
},
{
"URL": "https://academic.oup.com/bioinformatics/article-pdf/41/9/btaf491/64237229/btaf491.pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "286",
"original-title": [],
"prefix": "10.1093",
"published": {
"date-parts": [
[
2025,
9,
1
]
]
},
"published-online": {
"date-parts": [
[
2025,
9,
10
]
]
},
"published-other": {
"date-parts": [
[
2025,
9
]
]
},
"published-print": {
"date-parts": [
[
2025,
9,
1
]
]
},
"publisher": "Oxford University Press (OUP)",
"reference": [
{
"author": "Akiba",
"first-page": "2623",
"key": "2025091921101295900_btaf491-B1",
"year": "2019"
},
{
"DOI": "10.1038/s41564-021-00958-0",
"article-title": "Cellular host factors for SARS-CoV-2 infection",
"author": "Baggen",
"doi-asserted-by": "crossref",
"first-page": "1219",
"journal-title": "Nat Microbiol",
"key": "2025091921101295900_btaf491-B2",
"volume": "6",
"year": "2021"
},
{
"DOI": "10.15252/embj.2022111608",
"article-title": "Attenuation of SARS-CoV-2 replication and associated inflammation by concomitant targeting of viral and host cap 2′-o-ribose methyltransferases",
"author": "Bergant",
"doi-asserted-by": "crossref",
"first-page": "e111608",
"journal-title": "EMBO J",
"key": "2025091921101295900_btaf491-B3",
"volume": "41",
"year": "2022"
},
{
"DOI": "10.1093/nar/gkz203",
"article-title": "TriPepSVM: de novo prediction of RNA-binding proteins based on short amino acid motifs",
"author": "Bressin",
"doi-asserted-by": "crossref",
"first-page": "4406",
"journal-title": "Nucleic Acids Res",
"key": "2025091921101295900_btaf491-B4",
"volume": "47",
"year": "2019"
},
{
"article-title": "FlashAttention: fast and memory-efficient exact attention with io-awareness",
"author": "Dao",
"first-page": "16344",
"journal-title": "Adv Neural Inf Process Syst",
"key": "2025091921101295900_btaf491-B5",
"volume": "35",
"year": "2022"
},
{
"DOI": "10.1128/CMR.00102-15",
"article-title": "Approved antiviral drugs over the past 50 years",
"author": "De Clercq",
"doi-asserted-by": "crossref",
"first-page": "695",
"journal-title": "Clin Microbiol Rev",
"key": "2025091921101295900_btaf491-B6",
"volume": "29",
"year": "2016"
},
{
"first-page": "4171",
"key": "2025091921101295900_btaf491-B7"
},
{
"author": "Dhodapkar",
"key": "2025091921101295900_btaf491-B8",
"year": "2023"
},
{
"DOI": "10.1038/s42003-021-02095-0",
"article-title": "Genome-wide bioinformatic analyses predict key host and viral factors in SARS-CoV-2 pathogenesis",
"author": "Ferrarini",
"doi-asserted-by": "crossref",
"first-page": "590",
"journal-title": "Commun Biol",
"key": "2025091921101295900_btaf491-B9",
"volume": "4",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2023.04.003",
"article-title": "Structure of the endosomal commander complex linked to Ritscher-Schinzel syndrome",
"author": "Healy",
"doi-asserted-by": "crossref",
"first-page": "2219",
"journal-title": "Cell",
"key": "2025091921101295900_btaf491-B10",
"volume": "186",
"year": "2023"
},
{
"DOI": "10.1016/j.cels.2022.01.003",
"article-title": "Evolutionary velocity with protein language models predicts evolutionary dynamics of diverse proteins",
"author": "Hie",
"doi-asserted-by": "crossref",
"first-page": "274",
"journal-title": "Cell Syst",
"key": "2025091921101295900_btaf491-B11",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1093/nargab/lqad010",
"article-title": "A computational map of the human-SARS-CoV-2 protein–RNA interactome predicted at single-nucleotide resolution",
"author": "Horlacher",
"doi-asserted-by": "crossref",
"first-page": "lqad010",
"journal-title": "NAR Genom Bioinform",
"key": "2025091921101295900_btaf491-B12",
"volume": "5",
"year": "2023"
},
{
"key": "2025091921101295900_btaf491-B13"
},
{
"DOI": "10.1038/s41467-024-51074-6",
"article-title": "Multi-omics characterization of the monkeypox virus infection",
"author": "Huang",
"doi-asserted-by": "crossref",
"first-page": "6778",
"journal-title": "Nat Commun",
"key": "2025091921101295900_btaf491-B14",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1038/nrd.2017.162",
"article-title": "Host-directed therapies for bacterial and viral infections",
"author": "Kaufmann",
"doi-asserted-by": "crossref",
"first-page": "35",
"journal-title": "Nat Rev Drug Discov",
"key": "2025091921101295900_btaf491-B15",
"volume": "17",
"year": "2018"
},
{
"DOI": "10.1126/stke.2192004pl2",
"article-title": "Computational alanine scanning of protein–protein interfaces",
"author": "Kortemme",
"doi-asserted-by": "crossref",
"first-page": "pl2",
"journal-title": "Sci STKE",
"key": "2025091921101295900_btaf491-B16",
"volume": "2004",
"year": "2004"
},
{
"DOI": "10.1002/advs.202405058",
"article-title": "Machine learning early detection of SARS-CoV-2 high-risk variants",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "2405058",
"journal-title": "Adv Sci",
"key": "2025091921101295900_btaf491-B17",
"volume": "11",
"year": "2024"
},
{
"DOI": "10.1126/science.ade2574",
"article-title": "Evolutionary-scale prediction of atomic-level protein structure with a language model",
"author": "Lin",
"doi-asserted-by": "crossref",
"first-page": "1123",
"journal-title": "Science",
"key": "2025091921101295900_btaf491-B18",
"volume": "379",
"year": "2023"
},
{
"DOI": "10.1093/bib/bbaf276",
"article-title": "Generative prediction of real-world prevalent SARS-CoV-2 mutation with in silico virus evolution",
"author": "Liu",
"doi-asserted-by": "crossref",
"first-page": "bbaf276",
"journal-title": "Brief Bioinform",
"key": "2025091921101295900_btaf491-B19",
"volume": "26",
"year": "2025"
},
{
"DOI": "10.1093/nar/gkae241",
"article-title": "The EMBL-EBI job dispatcher sequence analysis tools framework in 2024",
"author": "Madeira",
"doi-asserted-by": "crossref",
"first-page": "W521",
"journal-title": "Nucleic Acids Res",
"key": "2025091921101295900_btaf491-B20",
"volume": "52",
"year": "2024"
},
{
"DOI": "10.1021/ja990935j",
"article-title": "Computational alanine scanning to probe protein–protein interactions: a novel approach to evaluate binding free energies",
"author": "Massova",
"doi-asserted-by": "crossref",
"first-page": "8133",
"journal-title": "J Am Chem Soc",
"key": "2025091921101295900_btaf491-B21",
"volume": "121",
"year": "1999"
},
{
"DOI": "10.1128/mbio.00009-23",
"article-title": "A virus-packageable CRISPR system identifies host dependency factors co-opted by multiple HIV-1 strains",
"author": "Montoya",
"doi-asserted-by": "crossref",
"first-page": "e00009",
"journal-title": "mBio",
"key": "2025091921101295900_btaf491-B22",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1016/S1367-5931(00)00206-4",
"article-title": "Combinatorial alanine-scanning",
"author": "Morrison",
"doi-asserted-by": "crossref",
"first-page": "302",
"journal-title": "Curr Opin Chem Biol",
"key": "2025091921101295900_btaf491-B23",
"volume": "5",
"year": "2001"
},
{
"DOI": "10.1073/pnas.2025581118",
"article-title": "Network medicine framework for identifying drug-repurposing opportunities for covid-19",
"author": "Morselli Gysi",
"doi-asserted-by": "crossref",
"first-page": "e2025581118",
"journal-title": "Proc Natl Acad Sci USA",
"key": "2025091921101295900_btaf491-B24",
"volume": "118",
"year": "2021"
},
{
"DOI": "10.1038/s41598-022-08073-8",
"article-title": "Computational identification of host genomic biomarkers highlighting their functions, pathways and regulators that influence SARS-CoV-2 infections and drug repurposing",
"author": "Mosharaf",
"doi-asserted-by": "crossref",
"first-page": "4279",
"journal-title": "Sci Rep",
"key": "2025091921101295900_btaf491-B25",
"volume": "12",
"year": "2022"
},
{
"author": "Rancati",
"key": "2025091921101295900_btaf491-B26"
},
{
"DOI": "10.1093/bib/bbae535",
"article-title": "Forecasting dominance of SARS-CoV-2 lineages by anomaly detection using deep autoencoders",
"author": "Rancati",
"doi-asserted-by": "crossref",
"first-page": "bbae535",
"journal-title": "Brief Bioinform",
"key": "2025091921101295900_btaf491-B27",
"volume": "25",
"year": "2024"
},
{
"DOI": "10.1093/bib/bbac456",
"article-title": "Discovery of host-directed modulators of virus infection by probing the SARS-CoV-2–host protein–protein interaction network",
"author": "Ravindran",
"doi-asserted-by": "crossref",
"first-page": "bbac456",
"journal-title": "Brief Bioinform",
"key": "2025091921101295900_btaf491-B28",
"volume": "23",
"year": "2022"
},
{
"DOI": "10.1038/s41588-022-01110-2",
"article-title": "Bidirectional genome-wide CRISPR screens reveal host factors regulating SARS-CoV-2, MERS-CoV and seasonal HCoVs",
"author": "Rebendenne",
"doi-asserted-by": "crossref",
"first-page": "1090",
"journal-title": "Nat Genet",
"key": "2025091921101295900_btaf491-B29",
"volume": "54",
"year": "2022"
},
{
"DOI": "10.1038/s41467-021-21770-8",
"article-title": "Identification of disease treatment mechanisms through the multiscale interactome",
"author": "Ruiz",
"doi-asserted-by": "crossref",
"first-page": "1796",
"journal-title": "Nat Commun",
"key": "2025091921101295900_btaf491-B30",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1038/s41598-022-15898-w",
"article-title": "SARS-CoV-2 potential drugs, drug targets, and biomarkers: a viral–host interaction network-based analysis",
"author": "Samy",
"doi-asserted-by": "crossref",
"first-page": "11934",
"journal-title": "Sci Rep",
"key": "2025091921101295900_btaf491-B31",
"volume": "12",
"year": "2022"
},
{
"DOI": "10.1038/s41586-018-0484-5",
"article-title": "An orthogonal proteomic survey uncovers novel Zika virus host factors",
"author": "Scaturro",
"doi-asserted-by": "crossref",
"first-page": "253",
"journal-title": "Nature",
"key": "2025091921101295900_btaf491-B32",
"volume": "561",
"year": "2018"
},
{
"DOI": "10.1038/s41467-024-51844-2",
"article-title": "Fine-tuning protein language models boosts predictions across diverse tasks",
"author": "Schmirler",
"doi-asserted-by": "crossref",
"first-page": "7407",
"journal-title": "Nat Commun",
"key": "2025091921101295900_btaf491-B33",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1093/nar/gkac194",
"article-title": "DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update)",
"author": "Sherman",
"doi-asserted-by": "crossref",
"first-page": "W216",
"journal-title": "Nucleic Acids Res",
"key": "2025091921101295900_btaf491-B34",
"volume": "50",
"year": "2022"
},
{
"DOI": "10.1038/nbt.3988",
"article-title": "MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets",
"author": "Steinegger",
"doi-asserted-by": "crossref",
"first-page": "1026",
"journal-title": "Nat Biotechnol",
"key": "2025091921101295900_btaf491-B35",
"volume": "35",
"year": "2017"
},
{
"DOI": "10.1038/s41586-021-03493-4",
"article-title": "Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV",
"author": "Stukalov",
"doi-asserted-by": "crossref",
"first-page": "246",
"journal-title": "Nature",
"key": "2025091921101295900_btaf491-B36",
"volume": "594",
"year": "2021"
},
{
"DOI": "10.1093/nar/gkac1000",
"article-title": "The string database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest",
"author": "Szklarczyk",
"doi-asserted-by": "crossref",
"first-page": "D638",
"journal-title": "Nucleic Acids Res",
"key": "2025091921101295900_btaf491-B37",
"volume": "51",
"year": "2023"
},
{
"DOI": "10.1038/s41586-023-06617-0",
"article-title": "Learning from prepandemic data to forecast viral escape",
"author": "Thadani",
"doi-asserted-by": "crossref",
"first-page": "818",
"journal-title": "Nature",
"key": "2025091921101295900_btaf491-B38",
"volume": "622",
"year": "2023"
},
{
"DOI": "10.1080/07391102.2021.1874532",
"article-title": "Structural similarity-based prediction of host factors associated with SARS-CoV-2 infection and pathogenesis",
"author": "Tiwari",
"doi-asserted-by": "crossref",
"first-page": "5868",
"journal-title": "J Biomol Struct Dyn",
"key": "2025091921101295900_btaf491-B39",
"volume": "40",
"year": "2022"
},
{
"DOI": "10.1093/nar/gkae1010",
"article-title": "Uniprot: the universal protein knowledgebase in 2025",
"author": "uni",
"doi-asserted-by": "crossref",
"first-page": "D609",
"journal-title": "Nucleic Acids Res",
"key": "2025091921101295900_btaf491-B40",
"volume": "53",
"year": "2025"
},
{
"DOI": "10.1093/nar/gkaa864",
"article-title": "Structural analysis of SARS-CoV-2 genome and predictions of the human interactome",
"author": "Vandelli",
"doi-asserted-by": "crossref",
"first-page": "11270",
"journal-title": "Nucleic Acids Res",
"key": "2025091921101295900_btaf491-B41",
"volume": "48",
"year": "2020"
},
{
"article-title": "Attention is all you need",
"author": "Vaswani",
"first-page": "5998",
"journal-title": "NIPS",
"key": "2025091921101295900_btaf491-B42"
},
{
"author": "Wang",
"key": "2025091921101295900_btaf491-B43",
"year": "2025"
},
{
"DOI": "10.1093/bib/bbae285",
"article-title": "Improved prediction of DNA and RNA binding proteins with deep learning models",
"author": "Wu",
"doi-asserted-by": "crossref",
"journal-title": "Brief Bioinform",
"key": "2025091921101295900_btaf491-B44",
"volume": "25",
"year": "2024"
},
{
"author": "Xiao",
"key": "2025091921101295900_btaf491-B45",
"year": "2025"
},
{
"DOI": "10.1016/j.immuni.2025.04.015",
"article-title": "Computationally designed proteins mimic antibody immune evasion in viral evolution",
"author": "Youssef",
"doi-asserted-by": "crossref",
"first-page": "1411",
"journal-title": "Immunity",
"key": "2025091921101295900_btaf491-B46",
"volume": "58",
"year": "2025"
},
{
"DOI": "10.1038/s41467-021-21213-4",
"article-title": "A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry",
"author": "Zhu",
"doi-asserted-by": "crossref",
"first-page": "961",
"journal-title": "Nat Commun",
"key": "2025091921101295900_btaf491-B47",
"volume": "12",
"year": "2021"
}
],
"reference-count": 47,
"references-count": 47,
"relation": {},
"resource": {
"primary": {
"URL": "https://academic.oup.com/bioinformatics/article/doi/10.1093/bioinformatics/btaf491/8250707"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "TransFactor—prediction of pro-viral SARS-CoV-2 host factors using a protein language model",
"type": "journal-article",
"volume": "41"
}