SARS-CoV-2 NSP6 reduces autophagosome size and affects viral replication via sigma-1 receptor
et al., Journal of Virology, doi:10.1128/jvi.00754-24, Nov 2024
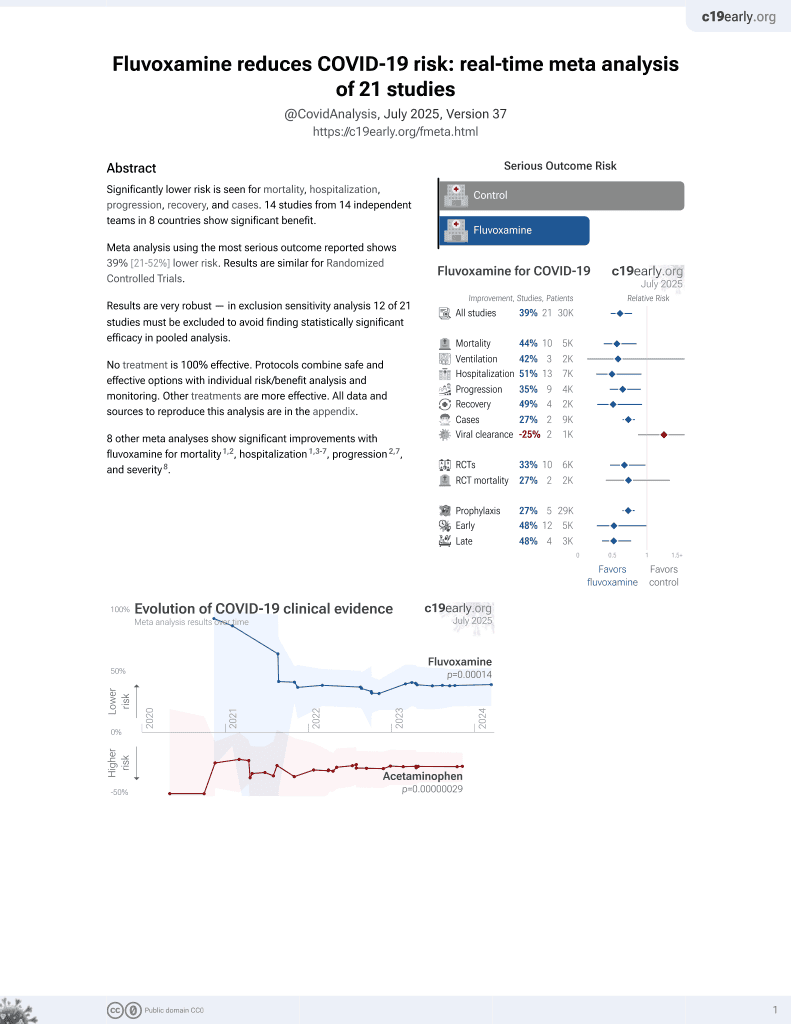
31st treatment shown to reduce risk in
November 2021, now with p = 0.00014 from 21 studies, recognized in 2 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
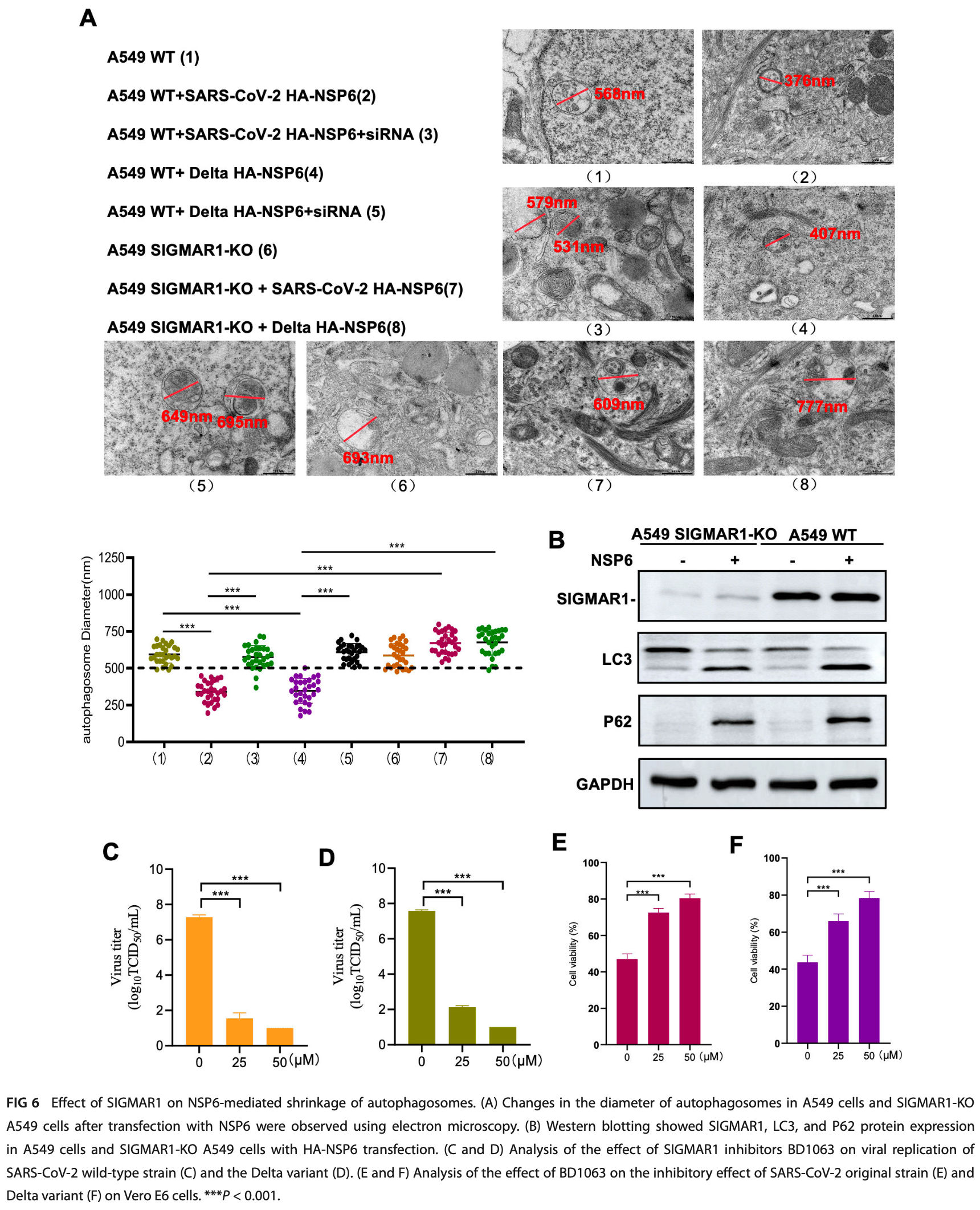
In vitro study showing that the NSP6 protein from SARS-CoV-2 wild-type and Delta variant strains induces smaller autophagosomes and affects lysosome function in A549 cells. Authors find that NSP6 activates autophagy through the Akt-mTOR-ULK1 pathway but blocks autophagosome-lysosome fusion, likely through endoplasmic reticulum-related pathways. The effects on autophagosome size depend on sigma-1 receptor (SIGMAR1), and SIGMAR1 knockout reverses the NSP6-induced autophagy abnormalities. In SIGMAR1 knockout cells, the SIGMAR1 inhibitor BD1063 reduces SARS-CoV-2 replication, suggesting SIGMAR1 may be a potential therapeutic target.
The results support the use of fluvoxamine (a SIGMAR1 agonist) for COVID-19. Fluvoxamine may help restore autophagic processes disrupted by NSP6, thereby reducing SARS-CoV-2 replication and improving host cellular defenses.
Zhang et al., 19 Nov 2024, USA, peer-reviewed, 13 authors.
Contact: lixiao06@mails.jlu.edu.cn, shangchao1290@126.com, liyq01@ccucm.edu.cn, liu820512@163.com.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
SARS-CoV-2 NSP6 reduces autophagosome size and affects viral replication via sigma-1 receptor
Journal of Virology, doi:10.1128/jvi.00754-24
Autophagy is a cellular self-defense mechanism by which cells can kill invading pathogenic microorganisms and increase the presentation of components of pathogens as antigens. Contrarily, pathogens can utilize autophagy to enhance their own replication. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) NSP6 can interact with ATPase proton pump component to inhibit lysosomal acidification, which was implicated in the autophagy process. However, research on how SARS-CoV-2 NSP6 affected autophagy, and its impact on virus replication is still lacking. Coronavirus NSP6 has been reported to promote coronavirus replication by limiting autophagosome expansion. However, this finding has not been confirmed in coronavirus disease 2019 . We investigated the effect of NSP6 protein on autophagosomes in differ ent mutant strains of SARS-CoV-2 and revealed that the size of autophagosomes was reduced by NSP6 of the wild-type and Delta variant of SARS-CoV-2. In addition, we found that SARS-CoV-2 NSP6 localized to the lysosome and had an inhibitory effect on the binding of autophagosomes to the lysosome, which blocked the autophagy flux; this may be related to endoplasmic reticulum (ER)-related pathways. We also found that sigma-1 receptor (SIGMAR1) knock out (KO) reversed NSP6-induced autophagosome abnormality and resisted SARS-CoV-2 infection, which responds to the fact that SIGMAR1 is likely to be used as a potential target for the treatment of SARS-CoV-2 infection. In summary, we have provided a preliminary explanation of the effects on autophagy of the SARS-CoV-2 NSP6 protein from the pre-autophagic and late stages, and also found that SIGMAR1 is likely to be used as a potential target for SARS-CoV-2 therapy to develop relevant drugs. IMPORTANCE We have provided a preliminary explanation of the effects on autoph agy of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) non-structure protein 6 from the pre-autophagic and late stages, and also found that sigma-1 receptor is likely to be used as a potential target for SARS-CoV-2 therapy to develop relevant drugs.
AUTHOR AFFILIATIONS
ADDITIONAL FILES The following material is available online.
Supplemental Material Supplemental figures (JVI00754-24-s0001.pdf). Figures S1 to S5.
References
Alizadeh, Glogowska, Thliveris, Kalantari, Shojaei et al., Autophagy modulates transforming growth factor beta 1 induced epithelial to mesenchymal transition in non-small cell lung cancer cells, Biochim Biophys Acta Mol Cell Res, doi:10.1016/j.bbamcr.2018.02.007
Alizadeh, Shojaei, Sepanjnia, Hashemi, Eftekharpour et al., Simultaneous detection of autophagy and epithelial to mesenchymal transition in the non-small cell lung cancer cells, Methods Mol Biol, doi:10.1007/7651_2017_84
Almási, Török, Valkusz, Tajti, Csonka et al., Sigma-1 receptor engages an antiinflammatory and antioxidant feedback loop mediated by peroxiredoxin in experimental colitis, Antioxidants (Basel), doi:10.3390/antiox9111081
Bajaj, Lotfi, Pal, Ronza, Di et al., Lysosome biogenesis in health and disease, J Neurochem, doi:10.1111/jnc.14564
Baliji, Cammer, Sobral, Baker, Detection of nonstructural protein 6 in murine coronavirus-infected cells and analysis of the transmembrane topology by using bioinformatics and molecular approaches, J Virol, doi:10.1128/JVI.00254-09
Cottam, Maier, Manifava, Vaux, Chandra-Schoenfelder et al., Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate, Autophagy, doi:10.4161/auto.7.11.16642
Cottam, Whelband, Wileman, Coronavirus NSP6 restricts autophagosome expansion, Autophagy, doi:10.4161/auto.29309
Deretic, Kimura, Timmins, Moseley, Chauhan et al., Immunologic manifestations of autophagy, J Clin Invest, doi:10.1172/JCI73945
Franco-Juárez, Coronel-Cruz, Hernández-Ochoa, Gómez-Manzo, Cárdenas-Rodríguez et al., TFEB; beyond its role as an autophagy and lysosomes regulator, Cells, doi:10.3390/cells11193153
Gordon, Jang, Bouhaddou, Xu, Obernier et al., A SARS-CoV-2 protein interaction map reveals targets for drug repurposing, Nature, doi:10.1038/s41586-020-2286-9
Hombach-Klonisch, Mehrpour, Shojaei, Harlos, Pitz et al., Glioblastoma and chemoresistance to alkylating agents: involvement of apoptosis, autophagy, and unfolded protein response, Pharmacol Ther, doi:10.1016/j.pharmthera.2017.10.017
Huang, Zhang, Han, Bai, Zhou et al., Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG, Autophagy, doi:10.1080/15548627.2022.2161728
Jia, Liu, Bao, Lv, Li et al., Delayed oseltamivir plus sirolimus treatment attenuates H1N1 virusinduced severe lung injury correlated with repressed NLRP3 inflamma some activation and inflammatory cell infiltration, PLoS Pathog, doi:10.1371/journal.ppat.1007428
Kistler, Huddleston, Bedford, Rapid and parallel adaptive mutations in spike S1 drive clade success in SARS-CoV-2, Cell Host Microbe, doi:10.1016/j.chom.2022.03.018
Kumar, Javed, Paddar, Eskelinen, Timmins et al., Mammalian hybrid prophagophore is a precursor to autophagosomes, Autophagy
Kumar, Kumar, Garg, Giri, An insight into SARS-CoV-2 membrane protein interaction with spike, envelope, and nucleocapsid proteins, J Biomol Struct Dyn, doi:10.1080/07391102.2021.2016490
Lee, Huang, Lee, Van De Leemput, Kane et al., Characterization of SARS-CoV-2 proteins reveals Orf6 pathogenicity, subcellular localization, host interactions and attenuation by Selinexor, Cell Biosci, doi:10.1186/s13578-021-00568-7
Lin, Li, Liu, Zhou, He et al., Nonstructural protein 6 of porcine epidemic diarrhea virus induces autophagy to promote viral replication via the PI3K/Akt/mTOR axis, Vet Microbiol, doi:10.1016/j.vetmic.2020.108684
Prentice, Jerome, Yoshimori, Mizushima, Denison, Coronavirus replication complex formation utilizes components of cellular autophagy, J Biol Chem, doi:10.1074/jbc.M306124200
Ramírez, Iwata, Park, Tsang, Kang et al., Folliculin interacting protein 1 maintains metabolic homeostasis during B cell development by modulating AMPK, mTORC1, and TFE3, J Immunol, doi:10.4049/jimmunol.1900395
Rashid, Dzakah, Wang, Tang, The ORF8 protein of SARS-CoV-2 induced endoplasmic reticulum stress and mediated immune evasion by antagonizing production of interferon beta, Virus Res, doi:10.1016/j.virusres.2021.198350
Reggiori, Monastyrska, Verheije, Calì, Ulasli et al., Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication, Cell Host Microbe, doi:10.1016/j.chom.2010.05.013
Shang, Zhuang, Zhang, Li, Zhu et al., Inhibition of autophagy suppresses SARS-CoV-2 replication and ameliorates pneumonia in hACE2 transgenic mice and xenografted human lung tissues, J Virol, doi:10.1128/JVI.01537-21
Shi, Nabar, Huang, Kehrl, SARS-Coronavirus Open Reading Frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes, Cell Death Discov, doi:10.1038/s41420-019-0181-7
Sun, Liu, Huang, Xu, Hu et al., SARS-CoV-2 non-structural protein 6 triggers NLRP3-dependent pyroptosis by targeting ATP6AP1, Cell Death Differ, doi:10.1038/s41418-021-00916-7
Wang, Liu, Luo, Cui, Zhang et al., Rational drug repositioning for coronavirusassociated diseases using directional mapping and sideeffect inference, iScience, doi:10.1016/j.isci.2022.105348
Wang, Wu, Yasui, Geva, Hayden et al., Nucleoporin POM121 signals TFEB-mediated autophagy via activation of SIGMAR1/sigma-1 receptor chaperone by pridopidine, Autophagy, doi:10.1080/15548627.2022.2063003
Xia, Cao, Xie, Zhang, Chen et al., Evasion of type I interferon by SARS-CoV-2, Cell Rep, doi:10.1016/j.celrep.2020.108234
Yordy, Iwasaki, Autophagy in the control and pathogenesis of viral infection, Curr Opin Virol, doi:10.1016/j.coviro.2011.05.016
Zhang, Li, Deng, Zhao, Huang et al., A thermostable mRNA vaccine against COVID-19, Cell, doi:10.1016/j.cell.2020.07.024
DOI record:
{
"DOI": "10.1128/jvi.00754-24",
"ISSN": [
"0022-538X",
"1098-5514"
],
"URL": "http://dx.doi.org/10.1128/jvi.00754-24",
"abstract": "<jats:title>ABSTRACT</jats:title>\n <jats:sec>\n <jats:title/>\n <jats:p>Autophagy is a cellular self-defense mechanism by which cells can kill invading pathogenic microorganisms and increase the presentation of components of pathogens as antigens. Contrarily, pathogens can utilize autophagy to enhance their own replication. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) NSP6 can interact with ATPase proton pump component to inhibit lysosomal acidification, which was implicated in the autophagy process. However, research on how SARS-CoV-2 NSP6 affected autophagy, and its impact on virus replication is still lacking. Coronavirus NSP6 has been reported to promote coronavirus replication by limiting autophagosome expansion. However, this finding has not been confirmed in coronavirus disease 2019 (COVID-19). We investigated the effect of NSP6 protein on autophagosomes in different mutant strains of SARS-CoV-2 and revealed that the size of autophagosomes was reduced by NSP6 of the wild-type and Delta variant of SARS-CoV-2. In addition, we found that SARS-CoV-2 NSP6 localized to the lysosome and had an inhibitory effect on the binding of autophagosomes to the lysosome, which blocked the autophagy flux; this may be related to endoplasmic reticulum (ER)-related pathways. We also found that sigma-1 receptor (SIGMAR1) knock out (KO) reversed NSP6-induced autophagosome abnormality and resisted SARS-CoV-2 infection, which responds to the fact that SIGMAR1 is likely to be used as a potential target for the treatment of SARS-CoV-2 infection. In summary, we have provided a preliminary explanation of the effects on autophagy of the SARS-CoV-2 NSP6 protein from the pre-autophagic and late stages, and also found that SIGMAR1 is likely to be used as a potential target for SARS-CoV-2 therapy to develop relevant drugs.</jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>IMPORTANCE</jats:title>\n <jats:p>We have provided a preliminary explanation of the effects on autophagy of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) non-structure protein 6 from the pre-autophagic and late stages, and also found that sigma-1 receptor is likely to be used as a potential target for SARS-CoV-2 therapy to develop relevant drugs.</jats:p>\n </jats:sec>",
"alternative-id": [
"10.1128/jvi.00754-24"
],
"assertion": [
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2024-04-26"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "2024-08-29"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 3,
"value": "2024-10-24"
}
],
"author": [
{
"ORCID": "http://orcid.org/0000-0003-3370-7021",
"affiliation": [
{
"name": "Changchun Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Changchun, China"
}
],
"authenticated-orcid": true,
"family": "Zhang",
"given": "Cuiling",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Changchun Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Changchun, China"
}
],
"family": "Jiang",
"given": "Qiwei",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Changchun Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Changchun, China"
}
],
"family": "Liu",
"given": "Zirui",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Changchun Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Changchun, China"
}
],
"family": "Li",
"given": "Nan",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Changchun Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Changchun, China"
}
],
"family": "Hao",
"given": "Zhuo",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Jiangxi Provincial Key Laboratory of Systems Biomedicine, Jiujiang University, Jiujiang, China"
}
],
"family": "Song",
"given": "Gaojie",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Neurosurgery, First Hospital of Jilin University, Changchun, China"
}
],
"family": "Li",
"given": "Dapeng",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Changchun Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Changchun, China"
}
],
"family": "Chen",
"given": "Minghua",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Chemistry, Northeastern University, Shenyang, China"
}
],
"family": "Lin",
"given": "Lisen",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0009-0006-6359-2615",
"affiliation": [
{
"name": "Changchun Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Changchun, China"
}
],
"authenticated-orcid": false,
"family": "Liu",
"given": "Yan",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0009-0004-0765-475X",
"affiliation": [
{
"name": "Changchun Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Changchun, China"
}
],
"authenticated-orcid": false,
"family": "Li",
"given": "Xiao",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0009-0008-4696-1575",
"affiliation": [
{
"name": "Changchun Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Changchun, China"
}
],
"authenticated-orcid": false,
"family": "Shang",
"given": "Chao",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-6795-6088",
"affiliation": [
{
"name": "Key Laboratory of Jilin Province for Traditional Chinese Medicine Prevention and Treatment of Infectious Diseases, College of Integrative Medicine, Changchun University of Chinese Medicine, Changchun, China"
}
],
"authenticated-orcid": false,
"family": "Li",
"given": "Yiquan",
"sequence": "additional"
}
],
"container-title": "Journal of Virology",
"container-title-short": "J Virol",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"journals.asm.org"
]
},
"created": {
"date-parts": [
[
2024,
10,
24
]
],
"date-time": "2024-10-24T12:47:53Z",
"timestamp": 1729774073000
},
"deposited": {
"date-parts": [
[
2024,
11,
19
]
],
"date-time": "2024-11-19T14:02:47Z",
"timestamp": 1732024967000
},
"editor": [
{
"affiliation": [],
"family": "Gallagher",
"given": "Tom",
"sequence": "additional"
}
],
"funder": [
{
"award": [
"20230508167RC"
],
"name": "Jilin Province Science and technology development plan youth growth science and technology"
},
{
"DOI": "10.13039/100007847",
"award": [
"YDZJ202201ZYTS257"
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/100007847",
"id-type": "DOI"
}
],
"name": "Natural Science Foundation of Jilin Province"
},
{
"DOI": "10.13039/501100001809",
"award": [
"82151221"
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100001809",
"id-type": "DOI"
}
],
"name": "MOST | National Natural Science Foundation of China"
}
],
"indexed": {
"date-parts": [
[
2024,
11,
19
]
],
"date-time": "2024-11-19T14:40:16Z",
"timestamp": 1732027216710,
"version": "3.28.0"
},
"is-referenced-by-count": 0,
"issue": "11",
"issued": {
"date-parts": [
[
2024,
11,
19
]
]
},
"journal-issue": {
"issue": "11",
"published-print": {
"date-parts": [
[
2024,
11,
19
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
11,
19
]
],
"date-time": "2024-11-19T00:00:00Z",
"timestamp": 1731974400000
}
},
{
"URL": "https://journals.asm.org/non-commercial-tdm-license",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
11,
19
]
],
"date-time": "2024-11-19T00:00:00Z",
"timestamp": 1731974400000
}
}
],
"link": [
{
"URL": "https://journals.asm.org/doi/pdf/10.1128/jvi.00754-24",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://journals.asm.org/doi/pdf/10.1128/jvi.00754-24",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "235",
"original-title": [],
"prefix": "10.1128",
"published": {
"date-parts": [
[
2024,
11,
19
]
]
},
"published-print": {
"date-parts": [
[
2024,
11,
19
]
]
},
"publisher": "American Society for Microbiology",
"reference": [
{
"DOI": "10.1016/j.pharmthera.2017.10.017",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_2_2"
},
{
"DOI": "10.1111/jnc.14564",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_3_2"
},
{
"DOI": "10.1016/j.bbamcr.2018.02.007",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_4_2"
},
{
"DOI": "10.1007/7651_2017_84",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_5_2"
},
{
"DOI": "10.1172/JCI73945",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_6_2"
},
{
"DOI": "10.1016/j.coviro.2011.05.016",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_7_2"
},
{
"DOI": "10.1371/journal.ppat.1007428",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_8_2"
},
{
"DOI": "10.1186/s13578-021-00568-7",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_9_2"
},
{
"DOI": "10.1038/s41418-021-00916-7",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_10_2"
},
{
"DOI": "10.1016/j.celrep.2020.108234",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_11_2"
},
{
"DOI": "10.1074/jbc.M306124200",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_12_2"
},
{
"DOI": "10.1016/j.chom.2010.05.013",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_13_2"
},
{
"DOI": "10.4161/auto.29309",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_14_2"
},
{
"DOI": "10.1038/s41586-020-2286-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_15_2"
},
{
"DOI": "10.1128/JVI.01537-21",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_16_2"
},
{
"DOI": "10.1016/j.cell.2020.07.024",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_17_2"
},
{
"DOI": "10.3390/cells11193153",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_18_2"
},
{
"DOI": "10.4049/jimmunol.1900395",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_19_2"
},
{
"DOI": "10.1016/j.virusres.2021.198350",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_20_2"
},
{
"DOI": "10.1080/07391102.2021.2016490",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_21_2"
},
{
"DOI": "10.1080/15548627.2022.2063003",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_22_2"
},
{
"DOI": "10.1016/j.isci.2022.105348",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_23_2"
},
{
"DOI": "10.3390/antiox9111081",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_24_2"
},
{
"DOI": "10.4161/auto.7.11.16642",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_25_2"
},
{
"DOI": "10.1128/JVI.00254-09",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_26_2"
},
{
"DOI": "10.1038/s41420-019-0181-7",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_27_2"
},
{
"DOI": "10.1016/j.chom.2022.03.018",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_28_2"
},
{
"DOI": "10.1016/j.vetmic.2020.108684",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_29_2"
},
{
"DOI": "10.1080/15548627.2017.1356975",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_30_2"
},
{
"DOI": "10.1080/15548627.2022.2161728",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_31_2"
}
],
"reference-count": 30,
"references-count": 30,
"relation": {},
"resource": {
"primary": {
"URL": "https://journals.asm.org/doi/10.1128/jvi.00754-24"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "SARS-CoV-2 NSP6 reduces autophagosome size and affects viral replication via sigma-1 receptor",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1128/asmj-crossmark-policy-page",
"volume": "98"
}