UDCA May Promote COVID-19 Recovery: A Cohort Study with AI-Aided Analysis
et al., medRxiv, doi:10.1101/2023.05.02.23289410, May 2023
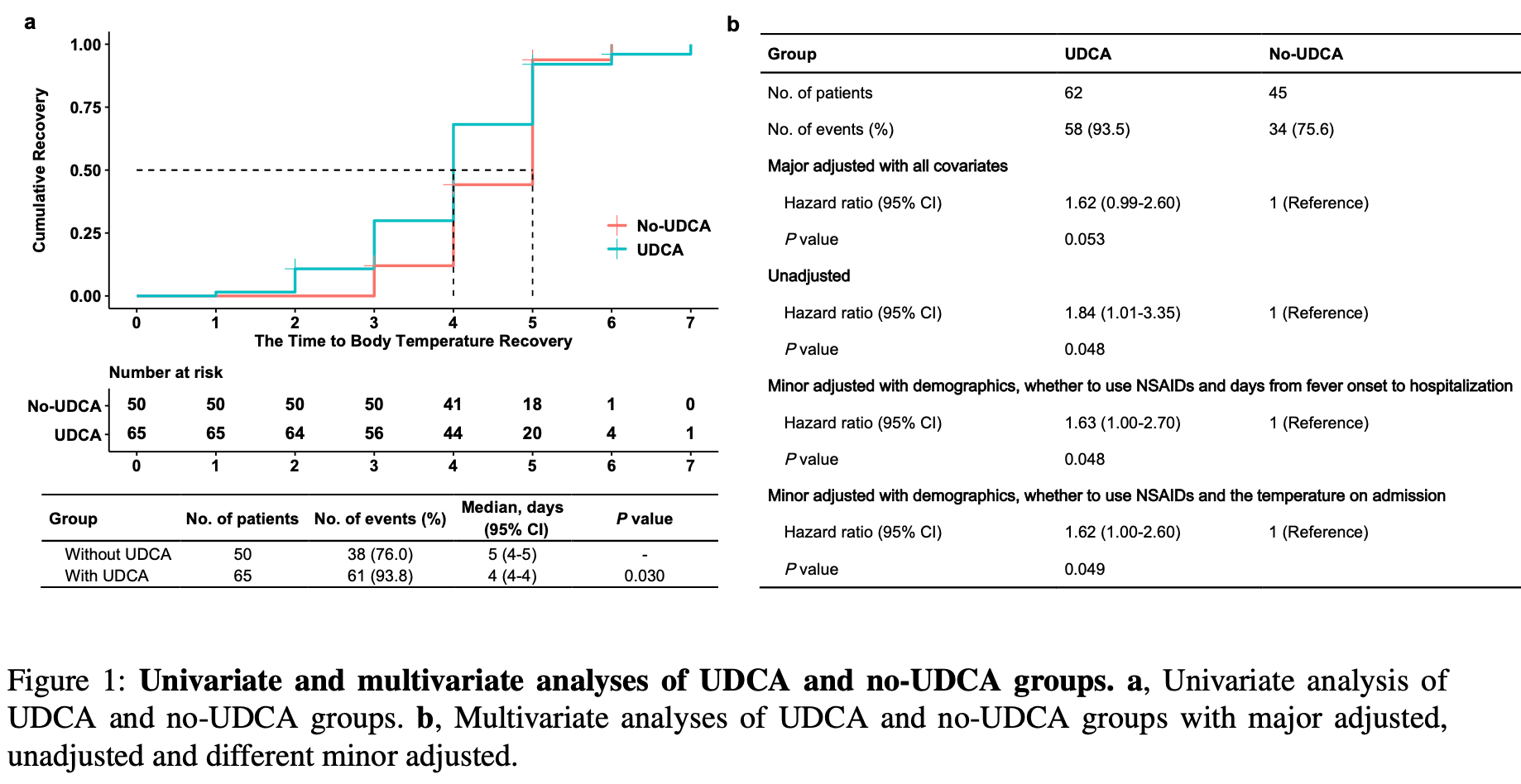
Retrospective 115 COVID-19 patients hospitalized during an Omicron outbreak in China, of which 65 received ursodeoxycholic acid (UDCA) treatment and 50 received standard care. It found that UDCA was associated with faster body temperature recovery, with a hazard ratio of 1.62 (95% CI 0.99-2.60, p=0.053) compared to standard care after adjusting for covariates. Patients receiving higher UDCA doses (≥300mg daily) had significantly faster recovery than the standard care group, with a hazard ratio of 1.82 (95% CI 1.07-3.10, p=0.028). To further analyze the exposure-response relationship, the authors developed an AI model called VirtualBody that accurately predicted individualized UDCA pharmacokinetic profiles. Additional analysis using VirtualBody-generated data found UDCA AUC, indicating total exposure over time, had a stronger correlation with clinical outcome than cumulative dose. Overall, the study suggests UDCA may shorten recovery time from COVID-19, especially at higher doses, warranting further investigation.
Standard of Care (SOC) for COVID-19 in the study country,
China, is average with moderate efficacy for approved treatments1.
|
risk of no recovery, 38.3% lower, HR 0.62, p = 0.05, treatment 62, control 45, inverted to make HR<1 favor treatment.
|
| Effect extraction follows pre-specified rules prioritizing more serious outcomes. Submit updates |
Yu et al., 4 May 2023, retrospective, China, preprint, 11 authors, study period 10 December, 2022 - 30 December, 2022.
UDCA May Promote COVID-19 Recovery: A Cohort Study with AI-Aided Analysis
doi:10.1101/2023.05.02.23289410
To investigate the impact of ursodeoxycholic acid (UDCA) treatment on the clinical outcome of mild and moderate COVID-19 cases, a retrospective analysis was conducted to evaluate the efficacy of UDCA on patients diagnosed with COVID-19 during the peak of the Omicron outbreak in China. This study presents promising results, demonstrating that UDCA significantly reduced the time to Body Temperature Recovery after admission and a higher daily dose seems to be associated with a better outcome without observed safety concerns. We also introduced VirtualBody, a physiologically plausible artificial neural network model, to generate an accurate depiction of the drug concentration-time curve individually, which represented the absorption, distribution, metabolism, and excretion of UDCA in each patient. It exhibits exceptional performance in modeling the complex PK-PD profile of UDCA, characterized by its endogenous and enterohepatic cycling properties, and further validates the effectiveness of UDCA as a treatment option from the drug exposure-response perspective. Our work highlights the potential of UDCA as a novel treatment option for periodic outbreaks of COVID-19 and introduces a new paradigm for PK-PD analysis in retrospective studies to provide evidence for optimal dosing strategies. The COVID-19 pandemic has caused an enormous global burden on public health, affecting civil societies, and hindering economic development 1 . In China, the number of COVID-19 infections skyrocketed in November 2022, following the government's active optimization and refinement of its COVID-19 response. The most prevalent strains were BA.5.2 (70.8%) and BF.7 (23.4%) 2,3 . Therefore, given the emergence of new variants and the persistent risk of periodic outbreaks during the era of Omicron, the need for accessible, oral therapeutic options to treat COVID-19 remains urgent, especially for individuals who have already been vaccinated 4-6 . Currently, only two oral antivirals, ritonavir-boosted nirmatrelvir and molnupiravir, have been approved for the treatment of mild or moderate COVID-19 under Emergency Use Authorization 7-10 . However, their availability is predominantly limited to individuals with affluent financial resources, and their efficacy against the Omicron (B.1.1.529) variant in vaccinated patients is not well established. Recent studies have explored the potential of UDCA, a classic FXR inhibitor, as a treatment option for COVID-
Extended Data Table 3
References
Bellemare, Autonomous navigation of stratospheric balloons using reinforcement learning, Nature
Brevini, FXR inhibition may protect from SARS-CoV-2 infection by reducing ACE2, Nature
Burkard, Von Eckardstein, Rentsch, Differentiated quantification of human bile acids in serum by high-performance liquid chromatography-tandem mass spectrometry, Journal of Chromatography B
Center, Control, The latest news and information about the epidemic situation of COVID-19
Danhof, Mechanism-based pharmacokinetic-pharmacodynamic (PK-PD modeling in translational drug, research
Degrave, Magnetic control of tokamak plasmas through deep reinforcement learning, Nature
Dilger, Effect of ursodeoxycholic acid on bile acid profiles and intestinal detoxification machinery in primary biliary cirrhosis and health, Journal of hepatology
Dyson, Novel therapeutic targets in primary biliary cirrhosis, Nature Reviews Gastroenterology & Hepatology
Fawzi, Discovering faster matrix multiplication algorithms with reinforcement learning, Nature
Gaziano, Actionable druggable genome-wide mendelian randomization identifies repurposing opportunities for COVID-19, Nature medicine
Geng, De novo molecular generation via connection-aware motif mining
Glatz, Trauner, Kerl, Müllegger, The importance of serum bile acid level analysis and treatment with ursodeoxycholic acid in intrahepatic cholestasis of pregnancy: a case series from central europe, Archives of dermatology
Grant, The prevalence of symptoms in 24,410 adults infected by the novel coronavirus (SARS-CoV-2; COVID-19): A systematic review and meta-analysis of 148 studies from 9 countries, PloS one
Hammond, Oral nirmatrelvir for high-risk, nonhospitalized adults with COVID-19, New England Journal of Medicine
Hu, Effect of common polymorphisms of the Farnesoid X receptor and bile acid transporters on the pharmacokinetics of ursodeoxycholic acid, Clinical and Experimental Pharmacology and Physiology
Jumper, Highly accurate protein structure prediction with alphafold, Nature
Li, Pivotal dose of pembrolizumab: A dose-finding strategy for immuno-oncology, Clinical Pharmacology & Therapeutics
Lu, Bender, Jin, Guan, Deep learning prediction of patient response time course from early data via neural-pharmacokinetic/pharmacodynamic modelling, Nature machine intelligence
Luo, A survey on model-based reinforcement learning
Lyashchenko, Systemic exposure to hydroxychloroquine and its relationship with outcome in severely ill COVID-19 patients in new york city, British Journal of Clinical Pharmacology
Mirhoseini, A graph placement methodology for fast chip design, Nature
Monteil, Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ace2, Cell
Mouton, Standardization of pharmacokinetic/pharmacodynamic (PK/PD) terminology for antiinfective drugs: an update, Journal of antimicrobial Chemotherapy
Prayitno, Bhat, Repurposing udca, an FXR inhibitor, to prevent SARS-Cov-2 infection
Reddy, Pharmacokinetics under the COVID-19 storm, British Journal of Clinical Pharmacology
Sheiner, Rosenberg, Marathe, Estimation of population characteristics of pharmacokinetic parameters from routine clinical data, Journal of Pharmacokinetics & Biopharmaceutics
Shi, Yu, Da, Chen, Zeng, Virtual-taobao: Virtualizing real-world online retail environment for reinforcement learning
Sinha, Efficacy and safety of molnupiravir in mild COVID-19 patients in india, Cureus
Sutton, Barto, Reinforcement learning -an introduction Adaptive computation and machine learning
Zhu, Analysis of human c24 bile acids metabolome in serum and urine based on enzyme digestion of conjugated bile acids and LC-MS determination of unconjugated bile acids, Analytical and bioanalytical chemistry
Zhu, Zhu, The effect of self-limiting on the prevention and control of the diffuse COVID-19 epidemic with delayed and temporal-spatial heterogeneous, BMC Infectious Diseases
Öztürk, Özgür, Ozkirimli, Deepdta: deep drug-target binding affinity prediction, Bioinformatics
DOI record:
{
"DOI": "10.1101/2023.05.02.23289410",
"URL": "http://dx.doi.org/10.1101/2023.05.02.23289410",
"abstract": "<jats:p>To investigate the impact of ursodeoxycholic acid (UDCA) treatment on the clinical outcome of mild and moderate COVID-19 cases, a retrospective analysis was conducted to evaluate the efficacy of UDCA on patients diagnosed with COVID-19 during the peak of the Omicron outbreak in China. This study presents promising results, demonstrating that UDCA significantly reduced the time to Body Temperature Recovery after admission and a higher daily dose seems to be associated with a better outcome without observed safety concerns. We also introduced VirtualBody, a physiologically plausible artificial neural network model, to generate an accurate depiction of the drug concentration-time curve individually, which represented the absorption, distribution, metabolism, and excretion of UDCA in each patient. It exhibits exceptional performance in modeling the complex PK-PD profile of UDCA, characterized by its endogenous and enterohepatic cycling properties, and further validates the effectiveness of UDCA as a treatment option from the drug exposure-response perspective. Our work highlights the potential of UDCA as a novel treatment option for periodic outbreaks of COVID-19 and introduces a new paradigm for PK-PD analysis in retrospective studies to provide evidence for optimal dosing strategies.</jats:p>",
"accepted": {
"date-parts": [
[
2023,
5,
4
]
]
},
"author": [
{
"affiliation": [],
"family": "Yu",
"given": "Yang",
"sequence": "first"
},
{
"affiliation": [],
"family": "Yu",
"given": "Guo",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Han",
"given": "Lu-Yao",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Li",
"given": "Jian",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zhang",
"given": "Zhi-Long",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Liu",
"given": "Tian-Shuo",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Li",
"given": "Ming-Feng",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zhan",
"given": "De-Chuan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Tang",
"given": "Shao-Qiu",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zhou",
"given": "Zhi-Hua",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wang",
"given": "Guang-Ji",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2023,
5,
4
]
],
"date-time": "2023-05-04T17:45:13Z",
"timestamp": 1683222313000
},
"deposited": {
"date-parts": [
[
2023,
5,
4
]
],
"date-time": "2023-05-04T17:45:13Z",
"timestamp": 1683222313000
},
"group-title": "Infectious Diseases (except HIV/AIDS)",
"indexed": {
"date-parts": [
[
2023,
5,
5
]
],
"date-time": "2023-05-05T04:41:45Z",
"timestamp": 1683261705750
},
"institution": [
{
"name": "medRxiv"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2023,
5,
4
]
]
},
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2023.05.02.23289410",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2023,
5,
4
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2023,
5,
4
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "http://medrxiv.org/lookup/doi/10.1101/2023.05.02.23289410"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subtitle": [],
"subtype": "preprint",
"title": "UDCA May Promote COVID-19 Recovery: A Cohort Study with AI-Aided Analysis",
"type": "posted-content"
}