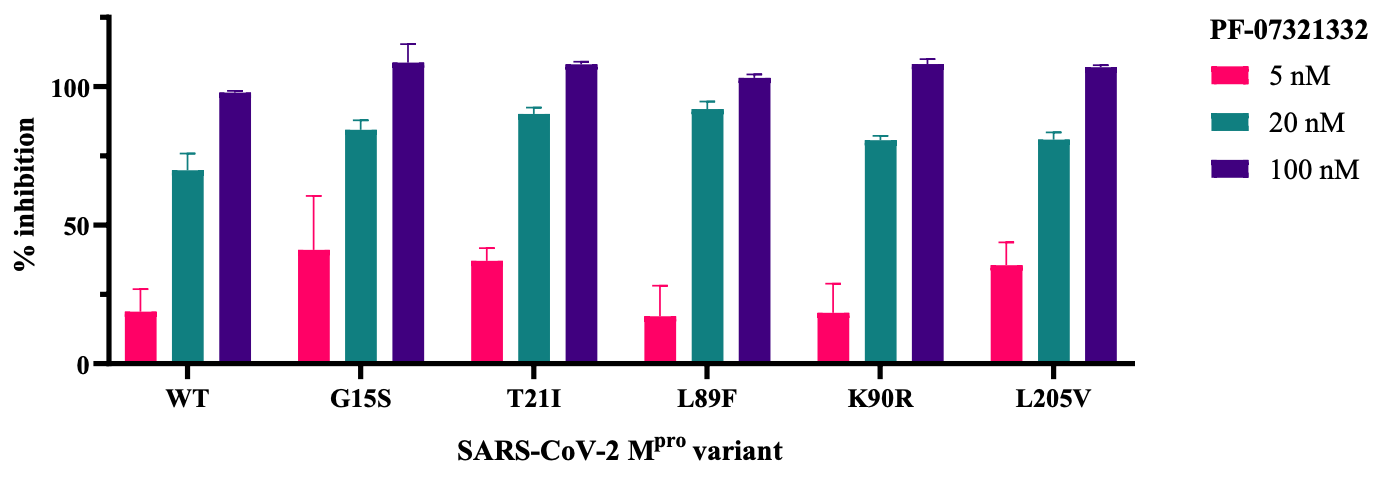
Main protease mutants of SARS-CoV-2 variants remain susceptible to PF-07321332
et al., bioRxiv, doi:10.1101/2021.11.28.4702264, Nov 2021
In vitro study showing that PF-07321332 maintains efficacy against variants C.37 lambda, B.1.1.318, B.1.2, B.1.351 beta, and P.2 zeta.
Ullrich et al., 30 Nov 2021, preprint, 4 authors.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Main protease mutants of SARS-CoV-2 variants remain susceptible to nirmatrelvir (PF-07321332)
doi:10.1101/2021.11.28.470226
The COVID-19 pandemic continues to be a public health threat. Multiple mutations in the spike protein of emerging variants of SARS-CoV-2 appear to impact on the effectiveness of available vaccines. Specific antiviral agents are keenly anticipated but their efficacy may also be compromised in emerging variants. One of the most attractive coronaviral drug targets is the main protease (M pro ). A promising M pro inhibitor of clinical relevance is the peptidomimetic PF-07321332. We expressed M pro of five SARS-CoV-2 lineages (C.37 Lambda, B.1.1.318, B.1.2, B.1.351 Beta, P.2 Zeta), each of which carries a strongly prevalent missense mutation (G15S, T21I, L89F, K90R, L205V). Enzyme kinetics showed that these M pro variants are similarly catalytically competent as the wildtype. We show that PF-07321332 has similar potency against the variants as against the wildtype. Our in vitro data suggest that the efficacy of specific M pro inhibitors such as PF-07321332 is not compromised in current COVID-19 variants. .
Competing Interest Declaration The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Author Information Sven Ullrich, ORCID: 0000-0003-4184-7024 Kasuni B. Ekanayake, ORCID: 0000-0002-3078-5895 Gottfried Otting, ORCID: 0000-0002-0563-0146 Christoph Nitsche, ORCID: 0000-0002-3704-2699
References
Agbowuro, Huston, Gamble, Tyndall, Proteases and protease inhibitors in infectious diseases, Med. Res. Rev, doi:10.1002/med.21475
Barrila, Gabelli, Bacha, Amzel, Freire, Mutation of Asn28 disrupts the dimerization and enzymatic activity of SARS 3CL pro, Biochemistry, doi:10.1021/bi1002585
Chen, Hu, Zhang, Chen, Chen et al., Mutation of Gly-11 on the dimer interface results in the complete crystallographic dimer dissociation of severe acute respiratory syndrome coronavirus 3C-like protease: crystal structure with molecular dynamics simulations, J. Biol. Chem, doi:10.1074/jbc.M705240200
Cheng, Chang, Chou, Mutation of Glu-166 blocks the substrateinduced dimerization of SARS coronavirus main protease, Biophys. J, doi:10.1016/j.bpj.2009.12.4272
Dong, Du, Gardner, An interactive web-based dashboard to track COVID-19 in real time, Lancet Infect. Dis, doi:10.1016/S1473-3099(20)30120-1
Dupont, Snell, Graham, Seow, Merrick et al., Neutralizing antibody activity in convalescent sera from infection in humans with SARS-CoV-2 and variants of concern, Nat. Microbiol, doi:10.1038/s41564-021-00974-0
Elbe, Buckland-Merrett, Data, disease and diplomacy: GISAID's innovative contribution to global health, Glob. Chall, doi:10.1002/gch2.1018
Fraternali, Lim, Shi, Mu, Song, Dynamically-driven enhancement of the catalytic machinery of the SARS 3C-like protease by the S284-T285-I286/A mutations on the extra domain, PLOS One, doi:10.1371/journal.pone.0101941
Hartenian, Nandakumar, Lari, Ly, Tucker et al., The molecular virology of coronaviruses, J. Biol. Chem, doi:10.1074/jbc.REV120.013930
Harvey, Carabelli, Jackson, Gupta, Thomson et al., None
He, He, Hong, Zhang, Wei, The challenges of COVID-19 Delta variant: prevention and vaccine development, MedComm, doi:10.1002/mco2.95
Hiscott, Alexandridi, Muscolini, Tassone, Palermo et al., The global impact of the coronavirus pandemic, Cytokine Growth Factor Rev
Hu, Zhang, Li, Wang, Chen et al., Two adjacent mutations on the dimer interface of SARS coronavirus 3C-like protease cause different conformational changes in crystal structure, Virology, doi:10.1016/j.virol.2009.03.034
Jin, Du, Xu, Deng, Liu et al., Structure of M pro from SARS-CoV-2 and discovery of its inhibitors, Nature, doi:10.1038/s41586-020-2223-y
Kim, Lee, Yang, Kim, Kim et al., The architecture of SARS-CoV-2 transcriptome, Cell, doi:10.1016/j.cell.2020.04.011
Latif, Mullen, Alkuzweny, Tsueng, Cano et al., info -ORF1a:G3278S mutation report
Latif, Mullen, Alkuzweny, Tsueng, Cano et al., info -ORF1a:K3353R mutation report
Latif, Mullen, Alkuzweny, Tsueng, Cano et al., info -ORF1a:L3352F mutation report
Latif, Mullen, Alkuzweny, Tsueng, Cano et al., info -ORF1a:L3468V mutation report
Latif, Mullen, Alkuzweny, Tsueng, Cano et al., info -ORF1a:T3284I mutation report
Latif, Mullen, Alkuzweny, Tsueng, Cano et al., info -lineage comparison
Le, Cramer, Chen, Mayhew, Evolution of the COVID-19 vaccine development landscape, Nat. Rev. Drug Discov, doi:10.1038/d41573-020-00151-8
Ludden, Reeve, Rambaut, Peacock, Coronavirus helicases: attractive and unique targets of antiviral drug-development and therapeutic patents, Expert Opin. Ther. Pat
Mahmoudvand, Shokri, Interactions between SARS coronavirus 2 papain-like protease and immune system: A potential drug target for the treatment of COVID-19, Scand. J. Immunol, doi:10.1111/sji.13044
Miyata, Miyazawa, Yasunaga, Two types of amino acid substitutions in protein evolution, J. Mol. Evol, doi:10.1007/bf01732340
Mullen, Tsueng, Latif, Alkuzweny, Cano et al., info -a standardized, open-source database of COVID-19 resources and epidemiology data
Owen, Allerton, Anderson, Aschenbrenner, Avery et al., An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19, Science, doi:10.1126/science.abl4784
Painter, Natchus, Cohen, Holman, Painter, Developing a direct acting, orally available antiviral agent in a pandemic: the evolution of molnupiravir as a potential treatment for COVID-19, Curr. Opin. Virol, doi:10.1016/j.coviro.2021.06.003
Rambaut, Holmes, O'toole, Hill, Mccrone et al., A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology, Nat. Microbiol, doi:10.1038/s41564-020-0770-5
Rut, Groborz, Zhang, Sun, Zmudzinski et al., SARS-CoV-2 M pro inhibitors and activity-based probes for patient-sample imaging, Nat. Chem. Biol, doi:10.1038/s41589-020-00689-z
Schechter, Berger, On the size of the active site in proteases. I. Papain, Biochem. Biophys. Res. Commun, doi:10.1016/s0006-291x(67)80055-x
Shi, Sivaraman, Song, Mechanism for controlling the dimer-monomer switch and coupling dimerization to catalysis of the severe acute respiratory syndrome coronavirus 3C-like protease, J. Virol, doi:10.1128/jvi.02680-07
Shi, Song, The catalysis of the SARS 3C-like protease is under extensive regulation by its extra domain, FEBS Journal, doi:10.1111/j.1742-4658.2006.05130
Subbarao, The success of SARS-CoV-2 vaccines and challenges ahead, Cell Host Microbe, doi:10.1016/j.chom.2021.06.016
Tao, Tzou, Nouhin, Gupta, De Oliveira et al., The biological and clinical significance of emerging SARS-CoV-2 variants, Nat. Rev. Genet, doi:10.1038/s41576-021-00408-x
Teijaro, Farber, COVID-19 vaccines: modes of immune activation and future challenges, Nat. Rev. Immunol, doi:10.1038/s41577-021-00526-x
Tregoning, Flight, Higham, Wang, Pierce, Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape, Nat. Rev. Immunol, doi:10.1038/s41577-021-00592-1
Ullrich, Nitsche, The SARS-CoV-2 main protease as drug target, Bioorg. Med. Chem. Lett, doi:10.1016/j.bmcl.2020.127377
Ullrich, Sasi, Mahawaththa, Ekanayake, Morewood et al., Challenges of short substrate . analogues as SARS-CoV-2 main protease inhibitors, Bioorg. Med. Chem. Lett, doi:10.1016/j.bmcl.2021.128333
Wensing, Van Maarseveen, Nijhuis, Fifteen years of HIV protease inhibitors: raising the barrier to resistance, Antivir. Res, doi:10.1016/j.antiviral.2009.10.003
Zeldovich, Chen, Shakhnovich, Protein stability imposes limits on organism complexity and speed of molecular evolution, Proc. Natl. Acad. Sci, doi:10.1073/pnas.0705366104
Zhang, Zeng, Gu, Li, Zheng et al., Progress and prospects on vaccine development against SARS-CoV-2, Vaccines, doi:10.3390/vaccines8020153
Zhu, Zhang, Wang, Li, Yang et al., A novel coronavirus from patients with pneumonia in China, 2019, N. Engl. J. Med, doi:10.1056/NEJMoa2001017
Zhukova, Blassel, Lemoine, Morel, Voznica et al., Origin, evolution and global spread of SARS-CoV-2, C. R. Biol, doi:10.5802/crbiol.29