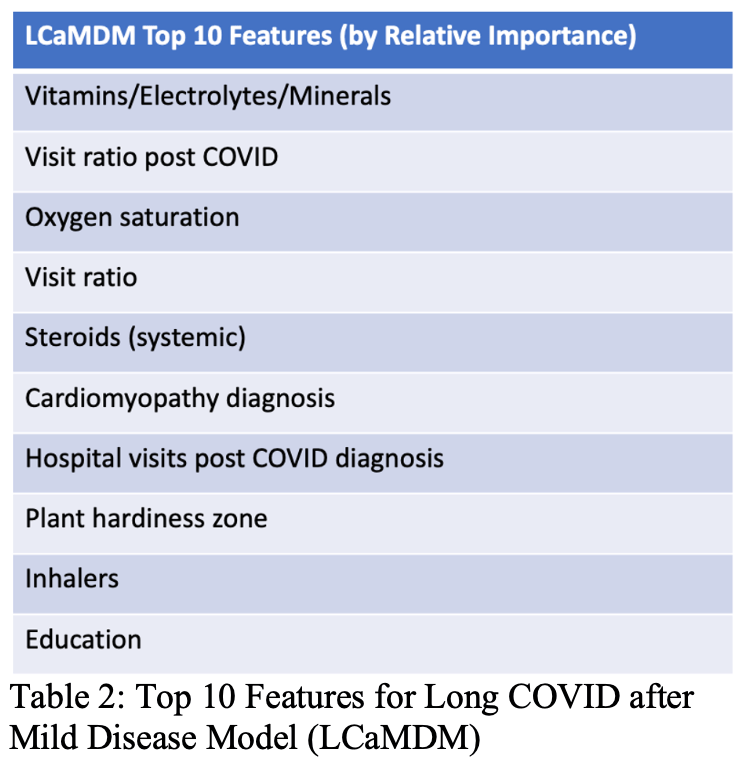
Prediction of Long COVID Based on Severity of Initial COVID-19 Infection: Differences in predictive feature sets between hospitalized versus non-hospitalized index infections
et al., medRxiv, doi:10.1101/2023.01.16.23284634, Jan 2023
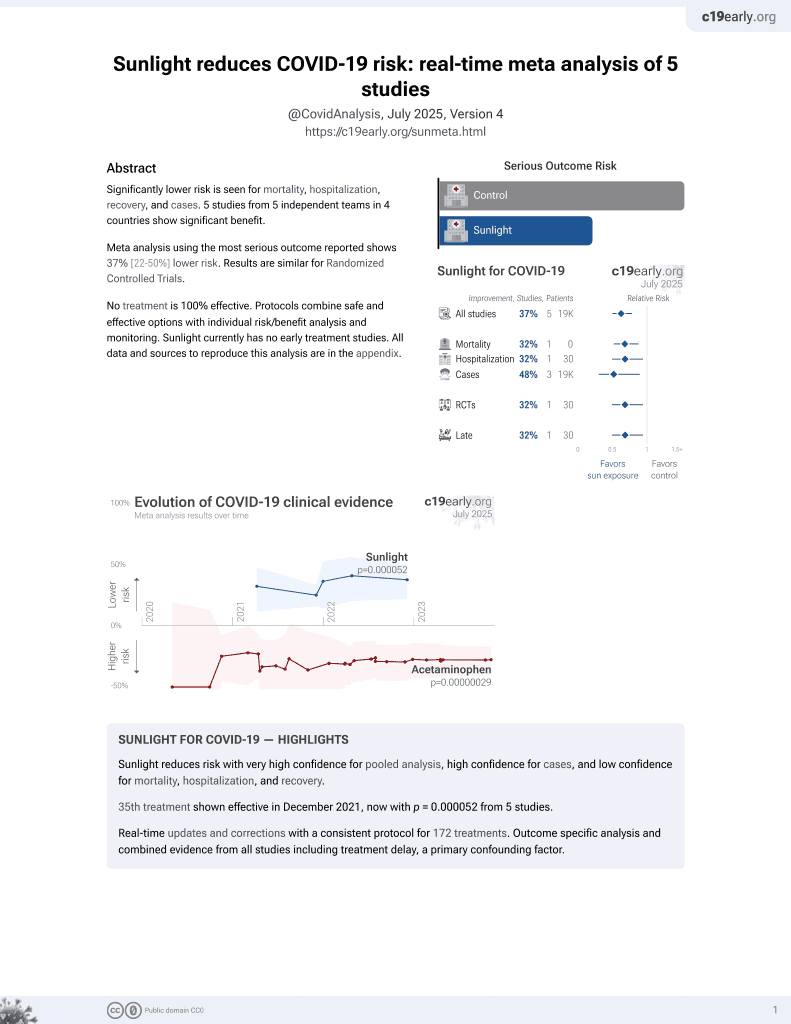
Sunlight for COVID-19
36th treatment shown to reduce risk in
December 2021, now with p = 0.000052 from 5 studies.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
N3C retrospective identifying plant hardiness zone as a predictive variable for long COVID. Authors note that this may be due to sunlight/climate affecting the risk of long COVID, and plan more detailed analysis in future work.
Socia et al., 18 Jan 2023, preprint, 5 authors.
Contact: robert.cockrell@med.uvm.edu.
Prediction of Long COVID Based on Severity of Initial COVID-19 Infection: Differences in predictive feature sets between hospitalized versus non-hospitalized index infections
doi:10.1101/2023.01.16.23284634
Long COVID is recognized as a significant consequence of SARS-COV2 infection. While the pathogenesis of Long COVID is still a subject of extensive investigation, there is considerable potential benefit in being able to predict which patients will develop Long COVID. We hypothesize that there would be distinct differences in the prediction of Long COVID based on the severity of the index infection, and use whether the index infection required hospitalization or not as a proxy for developing predictive models. We divide a large population of COVID patients drawn from the United States National Institutes of Health (NIH) National COVID Cohort Collaborative (N3C) Data Enclave Repository into two cohorts based on the severity of their initial COVID-19 illness and correspondingly trained two machine learning models: the Long COVID after Severe Disease Model (LCaSDM) and the Long COVID after Mild Disease Model (LCaMDM). The resulting models performed well on internal validation/testing, with a F1 score of 0.94 for the LCaSDM and 0.82 for the LCaMDM. There were distinct differences in the top 10 features used by each model, possibly reflecting the differences in type and amount of pathophysiological data between the hospitalized and non-hospitalized patients and/or reflecting different pathophysiological trajectories in the development of Long COVID. Of particular interest was the importance of Plant Hardiness Zone in the feature set for the LCaMDM, which may point to a role of climate and/or sunlight in the progression to Long COVID. Future work will involve a more detailed investigation of the potential role of climate and sunlight, as well as refinement of the predictive models as Long COVID becomes increasingly parsed into distinct clinical phenotypes.
Authors Statement: CC conceived the project strategy and developed environmental and demographic inputs. DS and DL implemented the ML model, and performed data cleaning, feature selection, ML model evaluation. SF parsed and interpreted clinical data features, aided in model analysis and extensively edited the manuscript. GA assisted with project design and manuscript development.
References: 1. National Institutes of Health (NIH). National Center for Advancing Translational Sciences (NCATS). National COVID Cohort Collaborative Data Enclave Repository. Bethesda,
References
Census, Bureau, P1: Total Population
Chen, Guestrin, Xgboost: A scalable tree boosting system
Johnson, Bulgarelli, Pollard, Horng, Celi et al., Mimic-iv. PhysioNet
Kachaner, Lemogne, Ranque, De Broucker, Meppiel, Somatic symptom disorder in patients with post-COVID-19 neurological symptoms: a preliminary report from the somatic study (Somatic Symptom Disorder Triggered by COVID-19), J Neurol Neurosurg Psychiatry
Perlis, Santillana, Ognyanova, Safarpour, Trujillo et al., Prevalence and correlates of long COVID symptoms among US adults, JAMA network open
Saunders, Sperling, Bendstrup, A new paradigm is needed to explain long COVID, Lancet Resp Med, doi:10.1016/S2213-2600(22)00501-X
Xie, Bowe, Al-Aly, Burdens of post-acute sequelae of COVID-19 by severity of acute infection, demographics and health status, Nature communications
Zhang, Zang, Xu, Zhang, Xu et al., Data-driven indentification of post-acute SARS-COV2 infection subphenotypes, Nature Medicine, doi:10.1038/s41591-022002116-3
DOI record:
{
"DOI": "10.1101/2023.01.16.23284634",
"URL": "http://dx.doi.org/10.1101/2023.01.16.23284634",
"abstract": "<jats:title>Abstract</jats:title><jats:p>Long COVID is recognized as a significant consequence of SARS-COV2 infection. While the pathogenesis of Long COVID is still a subject of extensive investigation, there is considerable potential benefit in being able to predict which patients will develop Long COVID. We hypothesize that there would be distinct differences in the prediction of Long COVID based on the severity of the index infection, and use whether the index infection required hospitalization or not as a proxy for developing predictive models. We divide a large population of COVID patients drawn from the United States National Institutes of Health (NIH) National COVID Cohort Collaborative (N3C) Data Enclave Repository into two cohorts based on the severity of their initial COVID-19 illness and correspondingly trained two machine learning models: the Long COVID after Severe Disease Model (LCaSDM) and the Long COVID after Mild Disease Model (LCaMDM). The resulting models performed well on internal validation/testing, with a F1 score of 0.94 for the LCaSDM and 0.82 for the LCaMDM. There were distinct differences in the top 10 features used by each model, possibly reflecting the differences in type and amount of pathophysiological data between the hospitalized and non-hospitalized patients and/or reflecting different pathophysiological trajectories in the development of Long COVID. Of particular interest was the importance of Plant Hardiness Zone in the feature set for the LCaMDM, which may point to a role of climate and/or sunlight in the progression to Long COVID. Future work will involve a more detailed investigation of the potential role of climate and sunlight, as well as refinement of the predictive models as Long COVID becomes increasingly parsed into distinct clinical phenotypes.</jats:p>",
"accepted": {
"date-parts": [
[
2023,
1,
18
]
]
},
"author": [
{
"affiliation": [],
"family": "Socia",
"given": "Damien",
"sequence": "first"
},
{
"affiliation": [],
"family": "Larie",
"given": "Dale",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Feuerwerker",
"given": "Sol",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-4549-9004",
"affiliation": [],
"authenticated-orcid": false,
"family": "An",
"given": "Gary",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Cockrell",
"given": "Chase",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2023,
1,
18
]
],
"date-time": "2023-01-18T17:10:18Z",
"timestamp": 1674061818000
},
"deposited": {
"date-parts": [
[
2023,
1,
20
]
],
"date-time": "2023-01-20T17:45:45Z",
"timestamp": 1674236745000
},
"group-title": "Infectious Diseases (except HIV/AIDS)",
"indexed": {
"date-parts": [
[
2023,
1,
21
]
],
"date-time": "2023-01-21T06:00:39Z",
"timestamp": 1674280839425
},
"institution": [
{
"name": "medRxiv"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2023,
1,
18
]
]
},
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2023.01.16.23284634",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2023,
1,
18
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2023,
1,
18
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference": [
{
"key": "2023012009450858000_2023.01.16.23284634v1.1",
"unstructured": "National Institutes of Health (NIH). National Center for Advancing Translational Sciences (NCATS). National COVID Cohort Collaborative Data Enclave Repository. Bethesda, Maryland: U.S. Department of Health and Human Services, National Institutes of Health, 2022."
},
{
"DOI": "10.1001/jamanetworkopen.2022.38804",
"article-title": "Prevalence and correlates of long COVID symptoms among US adults",
"doi-asserted-by": "crossref",
"first-page": "e2238804",
"issue": "10",
"journal-title": "JAMA network open",
"key": "2023012009450858000_2023.01.16.23284634v1.2",
"volume": "5",
"year": "2022"
},
{
"article-title": "Burdens of post-acute sequelae of COVID-19 by severity of acute infection, demographics and health status",
"first-page": "1",
"issue": "1",
"journal-title": "Nature communications",
"key": "2023012009450858000_2023.01.16.23284634v1.3",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1145/2939672.2939785",
"doi-asserted-by": "crossref",
"key": "2023012009450858000_2023.01.16.23284634v1.4",
"unstructured": "Chen, T. and C. Guestrin (2016). Xgboost: A scalable tree boosting system. Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining: 785–794."
},
{
"DOI": "10.1136/jnnp-2021-327899",
"doi-asserted-by": "publisher",
"key": "2023012009450858000_2023.01.16.23284634v1.5"
},
{
"key": "2023012009450858000_2023.01.16.23284634v1.6",
"unstructured": "Johnson, A. , Bulgarelli, L. , Pollard, T. , Horng, S. , Celi, L. A. , & Mark, R. (2020). Mimic-iv. PhysioNet. Available online at: https://physionet.org/content/mimiciv/1.0."
},
{
"key": "2023012009450858000_2023.01.16.23284634v1.7",
"unstructured": "U.S. Census Bureau. P1: Total Population. Available from: https://data.census.gov."
},
{
"key": "2023012009450858000_2023.01.16.23284634v1.8",
"unstructured": "PRISM Climate Group, Oregon State University, https://prism.oregonstate.edu, data created 4 Feb 2014, accessed 28 Oct. 2022."
},
{
"key": "2023012009450858000_2023.01.16.23284634v1.9",
"unstructured": "Google LLC. Google COVID-19 Community Mobility Reports”. [6/13/2021]. Available from: https://www.google.com/covid19/mobility."
},
{
"key": "2023012009450858000_2023.01.16.23284634v1.10",
"unstructured": "U.S. Department of Agriculture Economic Research Service. Educational attainment for adults age 25 and older for the U.S., States, and counties, 1970–2020. Available from: https://www.ers.usda.gov/data-products/county-level-data-sets/county-level-data-sets-download-data/"
},
{
"key": "2023012009450858000_2023.01.16.23284634v1.11",
"unstructured": "U.S. Department of Agriculture Economic Research Service. 2013 Rural-Urban Continuum Codes. Available from: https://www.ers.usda.gov/data-products/county-level-data-sets/county-level-data-sets-download-data/"
},
{
"DOI": "10.1038/s41591-022-02116-3",
"doi-asserted-by": "crossref",
"key": "2023012009450858000_2023.01.16.23284634v1.12",
"unstructured": "Zhang H , Zang C , Xu A , Zhang Y , Xu J , Bian J , Morozyuk D , Khullar D , Zhang Y , Nordvig AS , Schenck EJ , Shenkman EA , Rothman RL , Block JP , Lyman K , Weiner MG , Carton TW , Want F and Kaushal R. Data-driven indentification of post-acute SARS-COV2 infection subphenotypes. Nature Medicine (2022). DOI: https://doi.org/10.1038/s41591-022002116-3"
},
{
"DOI": "10.1016/S2213-2600(22)00501-X",
"doi-asserted-by": "crossref",
"key": "2023012009450858000_2023.01.16.23284634v1.13",
"unstructured": "Saunders C , Sperling S and Bendstrup E. A new paradigm is needed to explain long COVID. Lancet Resp Med (2023). Epub 5 January, 2023. DOI: https://doi.org/10.1016/S2213-2600(22)00501-X"
}
],
"reference-count": 13,
"references-count": 13,
"relation": {},
"resource": {
"primary": {
"URL": "http://medrxiv.org/lookup/doi/10.1101/2023.01.16.23284634"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subtitle": [],
"subtype": "preprint",
"title": "Prediction of Long COVID Based on Severity of Initial COVID-19 Infection: Differences in predictive feature sets between hospitalized versus non-hospitalized index infections",
"type": "posted-content"
}