Complex patterns of multimorbidity associated with severe COVID-19 and long COVID
et al., Communications Medicine, doi:10.1038/s43856-024-00506-x, Jul 2024
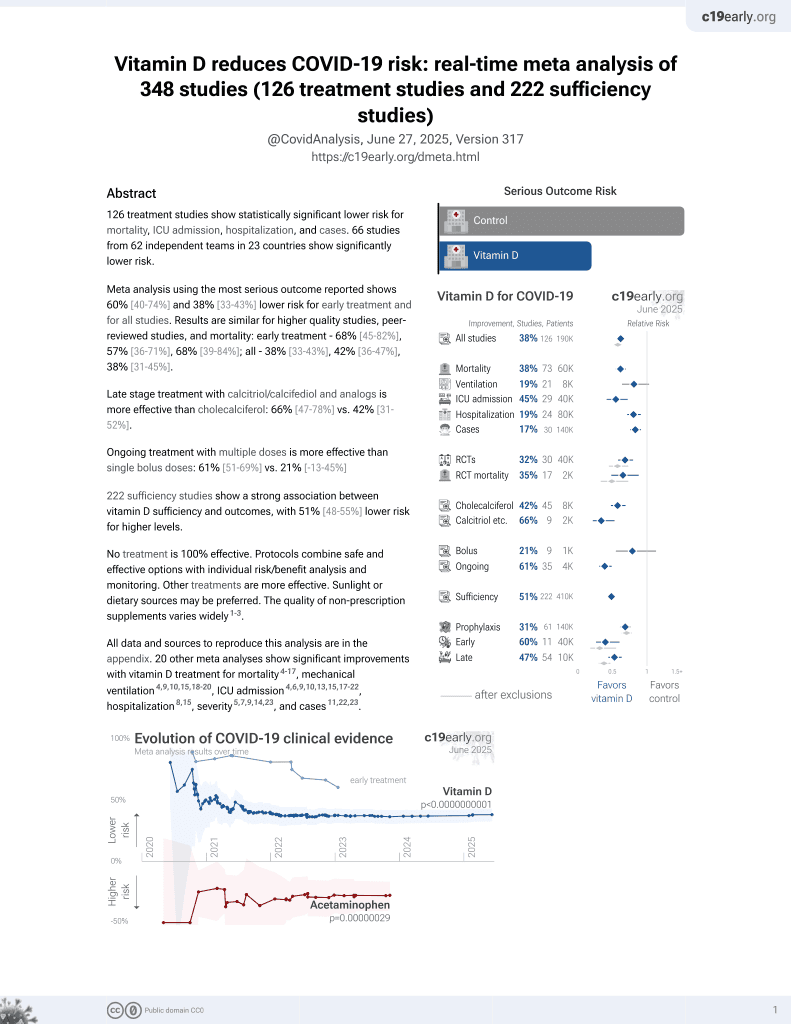
Vitamin D for COVID-19
8th treatment shown to reduce risk in
October 2020, now with p < 0.00000000001 from 135 studies, recognized in 18 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
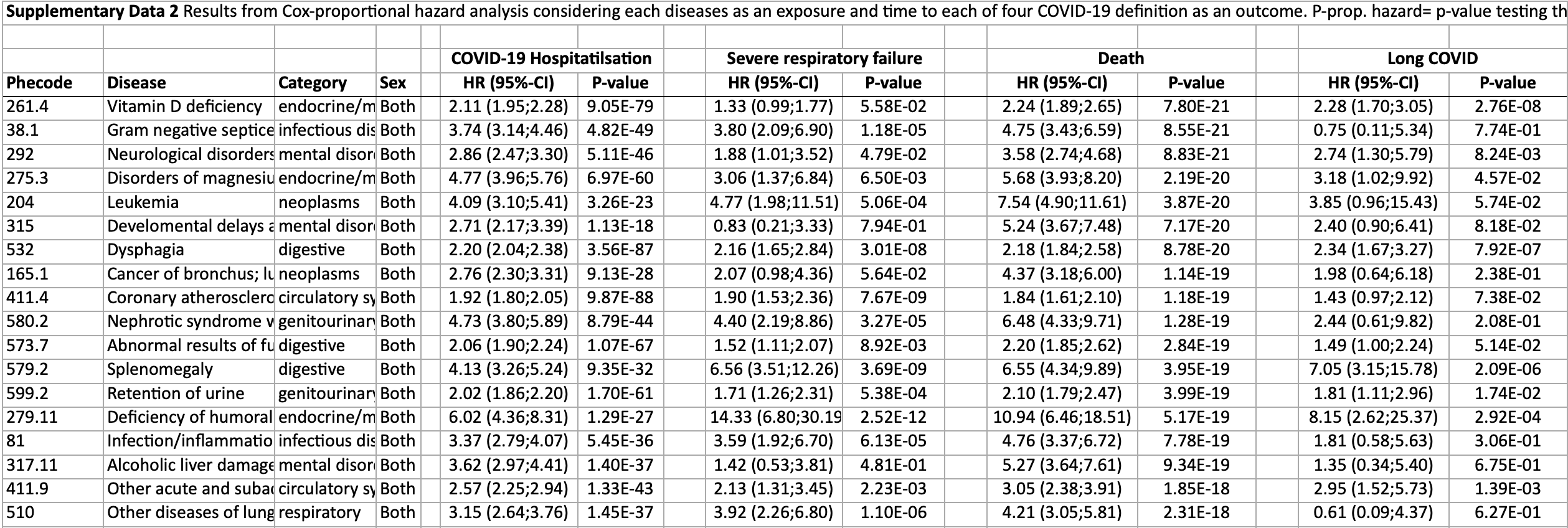
Retrospective 502,460 UK Biobank participants identifying many pre-existing conditions associated with increased risk of severe COVID-19 and long COVID, including vitamin D deficiency.
This is the 201st of 228 COVID-19 sufficiency studies for vitamin D, which collectively show higher levels reduce risk with p<0.0000000001.
Standard of Care (SOC) for COVID-19 in the study country,
the United Kingdom, is very poor with very low average efficacy for approved treatments1.
The United Kingdom focused on expensive high-profit treatments, approving only one low-cost early treatment, which required a prescription and had limited adoption. The high-cost prescription treatment strategy reduces the probability of early treatment due to access and cost barriers, and eliminates complementary and synergistic benefits seen with many low-cost treatments.
|
risk of death, 55.4% lower, HR 0.45, p < 0.001, cutoff 25nmol/L, inverted to make HR<1 favor high D levels (≥25nmol/L), Cox proportional hazards.
|
|
risk of hospitalization, 52.6% lower, HR 0.47, p < 0.001, cutoff 25nmol/L, inverted to make HR<1 favor high D levels (≥25nmol/L), Cox proportional hazards.
|
|
severe respiratory failure, 24.8% lower, HR 0.75, p = 0.05, cutoff 25nmol/L, inverted to make HR<1 favor high D levels (≥25nmol/L), Cox proportional hazards.
|
|
risk of long COVID, 56.1% lower, HR 0.44, p < 0.001, cutoff 25nmol/L, inverted to make HR<1 favor high D levels (≥25nmol/L), Cox proportional hazards.
|
| Effect extraction follows pre-specified rules prioritizing more serious outcomes. Submit updates |
Pietzner et al., 8 Jul 2024, retrospective, United Kingdom, peer-reviewed, 9 authors.
Contact: maik.pietzner@bih-charite.de, h.hemingway@ucl.ac.uk, claudia.langenberg@qmul.ac.uk.
Complex patterns of multimorbidity associated with severe COVID-19 and long COVID
Communications Medicine, doi:10.1038/s43856-024-00506-x
Background Early evidence that patients with (multiple) pre-existing diseases are at highest risk for severe COVID-19 has been instrumental in the pandemic to allocate critical care resources and later vaccination schemes. However, systematic studies exploring the breadth of medical diagnoses are scarce but may help to understand severe COVID-19 among patients at supposedly low risk. Methods We systematically harmonized >12 million primary care and hospitalisation health records from ~500,000 UK Biobank participants into 1448 collated disease terms to systematically identify diseases predisposing to severe COVID-19 (requiring hospitalisation or death) and its post-acute sequalae, Long COVID. Results Here we identify 679 diseases associated with an increased risk for severe COVID-19 (n = 672) and/or Long COVID (n = 72) that span almost all clinical specialties and are strongly enriched in clusters of cardio-respiratory and endocrine-renal diseases. For 57 diseases, we establish consistent evidence to predispose to severe COVID-19 based on survival and genetic susceptibility analyses. This includes a possible role of symptoms of malaise and fatigue as a so far largely overlooked risk factor for severe COVID-19. We finally observe partially opposing risk estimates at known risk loci for severe COVID-19 for etiologically related diseases, such as post-inflammatory pulmonary fibrosis or rheumatoid arthritis, possibly indicating a segregation of disease mechanisms. Conclusions Our results provide a unique reference that demonstrates how 1) complex cooccurrence of multipleincluding non-fatalconditions predispose to increased COVID-19 severity and 2) how incorporating the whole breadth of medical diagnosis can guide the interpretation of genetic risk loci. From the outset of the COVID-19 pandemic it was evident that underlying conditions were associated with both the risk of infection with SARS-CoV-2, the cause of COVID-19, and the risk of it being severe, based on the risk of hospitalisation, to ventilation and death 1 . Initial focus was on the small number of diseases known to put people at higher risk of other respiratory viral infections, such as influenza. The Center for Disease Control in the US and other national bodies published lists of diseases associated with COVID-19 and in the UK more than 1 million people were identified as
Competing interests The authors declare no competing interests.
Additional information Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s43856-024-00506-x. Correspondence and requests for materials should be addressed to Maik Pietzner, Harry Hemingway or Claudia Langenberg. Peer review information Communications Medicine thanks Marie Pigeyre and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available. Reprints
References
Barabasi, Albert, Emergence of scaling in random networks, Science
Bastarache, Using Phecodes for Research with the Electronic Health Record: From PheWAS to PheRS, Annu. Rev. Biomed. Data Sci
Booth, Population risk factors for severe disease and mortality in COVID-19: A global systematic review and meta-analysis, PloS One
Bulik-Sullivan, LD Score regression distinguishes confounding from polygenicity in genome-wide association studies, Nat. Genet
Bycroft, The UK Biobank resource with deep phenotyping and genomic data, Nature
Chimalakonda, Selectivity Profile of the Tyrosine Kinase 2 Inhibitor Deucravacitinib Compared with Janus Kinase 1/2/3 Inhibitors, Dermatol. Ther
Davis, Mccorkell, Vogel, Topol, Long COVID: major findings, mechanisms and recommendations, Nat. Rev. Microbiol
Denaxas, Mapping the Read2/CTV3 controlled clinical terminologies to Phecodes in UK Biobank primary care electronic health records: implementation and evaluation, Proc Am. Med. Inform. Assoc. Annu. Symp
Elyoussfi, Rane, Eyre, Warren, TYK2 as a novel therapeutic target in psoriasis, Expert Rev. Clin. Pharmacol
Fadista, Shared genetic etiology between idiopathic pulmonary fibrosis and COVID-19 severity, EBioMedicine
Giambartolomei, Bayesian test for colocalisation between pairs of genetic association studies using summary statistics, PLoS Genet
Hemani, The MR-Base platform supports systematic causal inference across the human phenome, eLife
Hoang, Baricitinib treatment resolves lower-airway macrophage inflammation and neutrophil recruitment in SARS-CoV-2-infected rhesus macaques, Cell
Kalil, Baricitinib plus Remdesivir for Hospitalized Adults with Covid-19, N. Engl. J. Med
Kompaniyets, Underlying Medical Conditions and Severe Illness Among 540,667 Adults Hospitalized With COVID-19, Prev. Chronic Dis
Kuan, Identifying and visualising multimorbidity and comorbidity patterns in patients in the English National Health Service: a population-based study, Lancet Digit. Health
Lammi, Genome-wide Association Study of Long COVID Authors
Letter, An EUA for baricitinib (Olumiant) for COVID-19, Med. Lett. Drugs Ther
Mbatchou, Computationally efficient whole-genome regression for quantitative and binary traits, Nat. Genet
Mckeigue, Rapid Epidemiological Analysis of Comorbidities and Treatments as risk factors for COVID-19 in Scotland (REACT-SCOT): A population-based case-control study, PLoS Med
Merad, Blish, Sallusto, Iwasaki, The immunology and immunopathology of COVID-19, Science
Pairo-Castineira, GWAS and meta-analysis identifies 49 genetic variants underlying critical COVID-19, Nature
Pairo-Castineira, Genetic mechanisms of critical illness in COVID-19, Nature
Strobl, Novel functions of tyrosine kinase 2 in the antiviral defense against murine cytomegalovirus, J. Immunol. Baltim. Md
Su, Epidemiology, clinical presentation, pathophysiology, and management of long COVID: an update, Mol. Psychiatry
Sudlow, UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age, PLoS Med
Thygesen, COVID-19 trajectories among 57 million adults in England: a cohort study using electronic health records, Lancet Digit. Health
Verbanck, Chen, Neale, Do, Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases, Nat. Genet
Verma, A MUC5B Gene Polymorphism, rs35705950-T, Confers Protective Effects Against COVID-19 Hospitalization but Not Severe Disease or Mortality, Am. J. Respir. Crit. Care Med
Verma, A Phenome-Wide Association Study of genes associated with COVID-19 severity reveals shared genetics with complex diseases in the Million Veteran Program, PLoS Genet
Wallace, A more accurate method for colocalisation analysis allowing for multiple causal variants, PLoS Genet
Wei, Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record, PLoS One
Williamson, Factors associated with COVID-19-related death using OpenSAFELY, Nature
Wu, Mapping ICD-10 and ICD-10-CM Codes to Phecodes: Workflow Development and Initial Evaluation, JMIR Med. Inform
Yavorska, Burgess, MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data, Int. J. Epidemiol
Zang, Data-driven analysis to understand long COVID using electronic health records from the RECOVER initiative, Nat. Commun
DOI record:
{
"DOI": "10.1038/s43856-024-00506-x",
"ISSN": [
"2730-664X"
],
"URL": "http://dx.doi.org/10.1038/s43856-024-00506-x",
"abstract": "<jats:title>Abstract</jats:title><jats:sec>\n <jats:title>Background</jats:title>\n <jats:p>Early evidence that patients with (multiple) pre-existing diseases are at highest risk for severe COVID-19 has been instrumental in the pandemic to allocate critical care resources and later vaccination schemes. However, systematic studies exploring the breadth of medical diagnoses are scarce but may help to understand severe COVID-19 among patients at supposedly low risk.</jats:p>\n </jats:sec><jats:sec>\n <jats:title>Methods</jats:title>\n <jats:p>We systematically harmonized >12 million primary care and hospitalisation health records from ~500,000 UK Biobank participants into 1448 collated disease terms to systematically identify diseases predisposing to severe COVID-19 (requiring hospitalisation or death) and its post-acute sequalae, Long COVID.</jats:p>\n </jats:sec><jats:sec>\n <jats:title>Results</jats:title>\n <jats:p>Here we identify 679 diseases associated with an increased risk for severe COVID-19 (<jats:italic>n</jats:italic> = 672) and/or Long COVID (<jats:italic>n</jats:italic> = 72) that span almost all clinical specialties and are strongly enriched in clusters of cardio-respiratory and endocrine-renal diseases. For 57 diseases, we establish consistent evidence to predispose to severe COVID-19 based on survival and genetic susceptibility analyses. This includes a possible role of symptoms of malaise and fatigue as a so far largely overlooked risk factor for severe COVID-19. We finally observe partially opposing risk estimates at known risk loci for severe COVID-19 for etiologically related diseases, such as post-inflammatory pulmonary fibrosis or rheumatoid arthritis, possibly indicating a segregation of disease mechanisms.</jats:p>\n </jats:sec><jats:sec>\n <jats:title>Conclusions</jats:title>\n <jats:p>Our results provide a unique reference that demonstrates how 1) complex co-occurrence of multiple – including non-fatal – conditions predispose to increased COVID-19 severity and 2) how incorporating the whole breadth of medical diagnosis can guide the interpretation of genetic risk loci.</jats:p>\n </jats:sec>",
"alternative-id": [
"506"
],
"article-number": "94",
"assertion": [
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Received",
"name": "received",
"order": 1,
"value": "9 November 2023"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "19 April 2024"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "First Online",
"name": "first_online",
"order": 3,
"value": "8 July 2024"
},
{
"group": {
"label": "Competing interests",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 1,
"value": "The authors declare no competing interests."
}
],
"author": [
{
"ORCID": "http://orcid.org/0000-0003-3437-9963",
"affiliation": [],
"authenticated-orcid": false,
"family": "Pietzner",
"given": "Maik",
"sequence": "first"
},
{
"affiliation": [],
"family": "Denaxas",
"given": "Spiros",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Yasmeen",
"given": "Summaira",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-9326-7227",
"affiliation": [],
"authenticated-orcid": false,
"family": "Ulmer",
"given": "Maria A.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-9510-5646",
"affiliation": [],
"authenticated-orcid": false,
"family": "Nakanishi",
"given": "Tomoko",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-4666-0923",
"affiliation": [],
"authenticated-orcid": false,
"family": "Arnold",
"given": "Matthias",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-2368-7322",
"affiliation": [],
"authenticated-orcid": false,
"family": "Kastenmüller",
"given": "Gabi",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-2279-0624",
"affiliation": [],
"authenticated-orcid": false,
"family": "Hemingway",
"given": "Harry",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5017-7344",
"affiliation": [],
"authenticated-orcid": false,
"family": "Langenberg",
"given": "Claudia",
"sequence": "additional"
}
],
"container-title": "Communications Medicine",
"container-title-short": "Commun Med",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"link.springer.com"
]
},
"created": {
"date-parts": [
[
2024,
7,
8
]
],
"date-time": "2024-07-08T10:11:29Z",
"timestamp": 1720433489000
},
"deposited": {
"date-parts": [
[
2024,
7,
8
]
],
"date-time": "2024-07-08T10:20:53Z",
"timestamp": 1720434053000
},
"indexed": {
"date-parts": [
[
2024,
7,
9
]
],
"date-time": "2024-07-09T00:24:37Z",
"timestamp": 1720484677038
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2024,
7,
8
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2024,
12
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
7,
8
]
],
"date-time": "2024-07-08T00:00:00Z",
"timestamp": 1720396800000
}
},
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
7,
8
]
],
"date-time": "2024-07-08T00:00:00Z",
"timestamp": 1720396800000
}
}
],
"link": [
{
"URL": "https://www.nature.com/articles/s43856-024-00506-x.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s43856-024-00506-x",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s43856-024-00506-x.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"prefix": "10.1038",
"published": {
"date-parts": [
[
2024,
7,
8
]
]
},
"published-online": {
"date-parts": [
[
2024,
7,
8
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference": [
{
"DOI": "10.1126/science.abm8108",
"author": "M Merad",
"doi-asserted-by": "publisher",
"first-page": "1122",
"journal-title": "Science",
"key": "506_CR1",
"unstructured": "Merad, M., Blish, C. A., Sallusto, F. & Iwasaki, A. The immunology and immunopathology of COVID-19. Science 375, 1122–1127 (2022).",
"volume": "375",
"year": "2022"
},
{
"key": "506_CR2",
"unstructured": "Clinically extremely vulnerable receive updated guidance in line with new national restrictions. GOV.UK https://www.gov.uk/government/news/clinically-extremely-vulnerable-receive-updated-guidance-in-line-with-new-national-restrictions (2024)."
},
{
"DOI": "10.5888/pcd18.210123",
"author": "L Kompaniyets",
"doi-asserted-by": "publisher",
"first-page": "E66",
"journal-title": "Prev. Chronic Dis.",
"key": "506_CR3",
"unstructured": "Kompaniyets, L. et al. Underlying Medical Conditions and Severe Illness Among 540,667 Adults Hospitalized With COVID-19, March 2020-March 2021. Prev. Chronic Dis. 18, E66 (2021).",
"volume": "18",
"year": "2021"
},
{
"DOI": "10.1371/journal.pone.0247461",
"author": "A Booth",
"doi-asserted-by": "publisher",
"first-page": "e0247461",
"journal-title": "PloS One",
"key": "506_CR4",
"unstructured": "Booth, A. et al. Population risk factors for severe disease and mortality in COVID-19: A global systematic review and meta-analysis. PloS One 16, e0247461 (2021).",
"volume": "16",
"year": "2021"
},
{
"DOI": "10.1038/s41586-020-2521-4",
"author": "EJ Williamson",
"doi-asserted-by": "publisher",
"first-page": "430",
"journal-title": "Nature",
"key": "506_CR5",
"unstructured": "Williamson, E. J. et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature 584, 430–436 (2020).",
"volume": "584",
"year": "2020"
},
{
"DOI": "10.1371/journal.pmed.1003374",
"author": "PM McKeigue",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "PLoS Med.",
"key": "506_CR6",
"unstructured": "McKeigue, P. M. et al. Rapid Epidemiological Analysis of Comorbidities and Treatments as risk factors for COVID-19 in Scotland (REACT-SCOT): A population-based case-control study. PLoS Med. 17, 1–17 (2020).",
"volume": "17",
"year": "2020"
},
{
"DOI": "10.1038/s41586-023-06034-3",
"author": "E Pairo-Castineira",
"doi-asserted-by": "publisher",
"first-page": "764",
"journal-title": "Nature",
"key": "506_CR7",
"unstructured": "Pairo-Castineira, E. et al. GWAS and meta-analysis identifies 49 genetic variants underlying critical COVID-19. Nature 617, 764–768 (2023).",
"volume": "617",
"year": "2023"
},
{
"key": "506_CR8",
"unstructured": "COVID-19 Host Genetics Initiative. A second update on mapping the human genetic architecture of COVID-19. Nature 621, E7–E26 (2023)."
},
{
"DOI": "10.1371/journal.pmed.1001779",
"author": "C Sudlow",
"doi-asserted-by": "publisher",
"first-page": "e1001779",
"journal-title": "PLoS Med",
"key": "506_CR9",
"unstructured": "Sudlow, C. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 12, e1001779 (2015).",
"volume": "12",
"year": "2015"
},
{
"DOI": "10.1371/journal.pone.0175508",
"author": "W-Q Wei",
"doi-asserted-by": "publisher",
"first-page": "e0175508",
"journal-title": "PLoS One",
"key": "506_CR10",
"unstructured": "Wei, W.-Q. et al. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS One 12, e0175508 (2017).",
"volume": "12",
"year": "2017"
},
{
"DOI": "10.1038/s41586-018-0579-z",
"author": "C Bycroft",
"doi-asserted-by": "publisher",
"first-page": "203",
"journal-title": "Nature",
"key": "506_CR11",
"unstructured": "Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).",
"volume": "562",
"year": "2018"
},
{
"DOI": "10.1016/S2589-7500(22)00091-7",
"author": "JH Thygesen",
"doi-asserted-by": "publisher",
"first-page": "e542",
"journal-title": "Lancet Digit. Health",
"key": "506_CR12",
"unstructured": "Thygesen, J. H. et al. COVID-19 trajectories among 57 million adults in England: a cohort study using electronic health records. Lancet Digit. Health 4, e542–e557 (2022).",
"volume": "4",
"year": "2022"
},
{
"key": "506_CR13",
"unstructured": "Lammi, V. et al. Genome-wide Association Study of Long COVID Authors. 1–25 (2023)."
},
{
"DOI": "10.2196/14325",
"author": "P Wu",
"doi-asserted-by": "publisher",
"first-page": "e14325",
"journal-title": "JMIR Med. Inform.",
"key": "506_CR14",
"unstructured": "Wu, P. et al. Mapping ICD-10 and ICD-10-CM Codes to Phecodes: Workflow Development and Initial Evaluation. JMIR Med. Inform. 7, e14325 (2019).",
"volume": "7",
"year": "2019"
},
{
"author": "S Denaxas",
"first-page": "362",
"journal-title": "Proc Am. Med. Inform. Assoc. Annu. Symp.",
"key": "506_CR15",
"unstructured": "Denaxas, S. Mapping the Read2/CTV3 controlled clinical terminologies to Phecodes in UK Biobank primary care electronic health records: implementation and evaluation. Proc Am. Med. Inform. Assoc. Annu. Symp. 2021, 362–371 (2021).",
"volume": "2021",
"year": "2021"
},
{
"DOI": "10.1146/annurev-biodatasci-122320-112352",
"author": "L Bastarache",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Annu. Rev. Biomed. Data Sci.",
"key": "506_CR16",
"unstructured": "Bastarache, L. Using Phecodes for Research with the Electronic Health Record: From PheWAS to PheRS. Annu. Rev. Biomed. Data Sci. 4, 1–19 (2021).",
"volume": "4",
"year": "2021"
},
{
"DOI": "10.1038/s41467-023-37653-z",
"author": "C Zang",
"doi-asserted-by": "publisher",
"journal-title": "Nat. Commun.",
"key": "506_CR17",
"unstructured": "Zang, C. et al. Data-driven analysis to understand long COVID using electronic health records from the RECOVER initiative. Nat. Commun. 14, 1948 (2023).",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1016/S2589-7500(22)00187-X",
"author": "V Kuan",
"doi-asserted-by": "publisher",
"first-page": "e16",
"journal-title": "Lancet Digit. Health",
"key": "506_CR18",
"unstructured": "Kuan, V. et al. Identifying and visualising multimorbidity and comorbidity patterns in patients in the English National Health Service: a population-based study. Lancet Digit. Health 5, e16–e27 (2023).",
"volume": "5",
"year": "2023"
},
{
"DOI": "10.1126/science.286.5439.509",
"author": "A Barabasi",
"doi-asserted-by": "publisher",
"first-page": "509",
"journal-title": "Science",
"key": "506_CR19",
"unstructured": "Barabasi, A. & Albert, R. Emergence of scaling in random networks. Science 286, 509–512 (1999).",
"volume": "286",
"year": "1999"
},
{
"DOI": "10.1038/s41588-021-00870-7",
"author": "J Mbatchou",
"doi-asserted-by": "publisher",
"first-page": "1097",
"journal-title": "Nat. Genet.",
"key": "506_CR20",
"unstructured": "Mbatchou, J. et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 53, 1097–1103 (2021).",
"volume": "53",
"year": "2021"
},
{
"DOI": "10.1038/ng.3211",
"author": "BK Bulik-Sullivan",
"doi-asserted-by": "publisher",
"first-page": "291",
"journal-title": "Nat. Genet.",
"key": "506_CR21",
"unstructured": "Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).",
"volume": "47",
"year": "2015"
},
{
"DOI": "10.1038/s41588-018-0099-7",
"author": "M Verbanck",
"doi-asserted-by": "publisher",
"first-page": "693",
"journal-title": "Nat. Genet.",
"key": "506_CR22",
"unstructured": "Verbanck, M., Chen, C.-Y., Neale, B. & Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698 (2018).",
"volume": "50",
"year": "2018"
},
{
"DOI": "10.1038/s41586-021-03767-x",
"author": "COVID-19 Host Genetics Initiative.",
"doi-asserted-by": "publisher",
"first-page": "472",
"journal-title": "Nature",
"key": "506_CR23",
"unstructured": "COVID-19 Host Genetics Initiative. Mapping the human genetic architecture of COVID-19. Nature 600, 472–477 (2021).",
"volume": "600",
"year": "2021"
},
{
"DOI": "10.1093/ije/dyx034",
"author": "OO Yavorska",
"doi-asserted-by": "publisher",
"first-page": "1734",
"journal-title": "Int. J. Epidemiol.",
"key": "506_CR24",
"unstructured": "Yavorska, O. O. & Burgess, S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int. J. Epidemiol. 46, 1734–1739 (2017).",
"volume": "46",
"year": "2017"
},
{
"DOI": "10.7554/eLife.34408",
"author": "G Hemani",
"doi-asserted-by": "publisher",
"first-page": "e34408",
"journal-title": "eLife",
"key": "506_CR25",
"unstructured": "Hemani, G. et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 7, e34408 (2018).",
"volume": "7",
"year": "2018"
},
{
"DOI": "10.1371/journal.pgen.1004383",
"author": "C Giambartolomei",
"doi-asserted-by": "publisher",
"first-page": "e1004383",
"journal-title": "PLoS Genet",
"key": "506_CR26",
"unstructured": "Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 10, e1004383 (2014).",
"volume": "10",
"year": "2014"
},
{
"DOI": "10.1371/journal.pgen.1009440",
"author": "C Wallace",
"doi-asserted-by": "publisher",
"first-page": "e1009440",
"journal-title": "PLoS Genet.",
"key": "506_CR27",
"unstructured": "Wallace, C. A more accurate method for colocalisation analysis allowing for multiple causal variants. PLoS Genet. 17, e1009440 (2021).",
"volume": "17",
"year": "2021"
},
{
"DOI": "10.1038/s41380-023-02171-3",
"author": "S Su",
"doi-asserted-by": "publisher",
"first-page": "4056",
"journal-title": "Mol. Psychiatry",
"key": "506_CR28",
"unstructured": "Su, S. et al. Epidemiology, clinical presentation, pathophysiology, and management of long COVID: an update. Mol. Psychiatry 28, 4056–4069 (2023).",
"volume": "28",
"year": "2023"
},
{
"DOI": "10.1038/s41586-020-03065-y",
"author": "E Pairo-Castineira",
"doi-asserted-by": "publisher",
"first-page": "92",
"journal-title": "Nature",
"key": "506_CR29",
"unstructured": "Pairo-Castineira, E. et al. Genetic mechanisms of critical illness in COVID-19. Nature 591, 92–98 (2021).",
"volume": "591",
"year": "2021"
},
{
"author": "TM Letter",
"first-page": "202",
"journal-title": "Med. Lett. Drugs Ther.",
"key": "506_CR30",
"unstructured": "Letter, T. M. An EUA for baricitinib (Olumiant) for COVID-19. Med. Lett. Drugs Ther. 62, 202–203 (2020).",
"volume": "62",
"year": "2020"
},
{
"DOI": "10.1056/NEJMoa2031994",
"author": "AC Kalil",
"doi-asserted-by": "publisher",
"first-page": "795",
"journal-title": "N. Engl. J. Med.",
"key": "506_CR31",
"unstructured": "Kalil, A. C. et al. Baricitinib plus Remdesivir for Hospitalized Adults with Covid-19. N. Engl. J. Med. 384, 795–807 (2021).",
"volume": "384",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.11.007",
"author": "TN Hoang",
"doi-asserted-by": "publisher",
"first-page": "460",
"journal-title": "Cell",
"key": "506_CR32",
"unstructured": "Hoang, T. N. et al. Baricitinib treatment resolves lower-airway macrophage inflammation and neutrophil recruitment in SARS-CoV-2-infected rhesus macaques. Cell 184, 460–475.e21 (2021).",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1007/s13555-021-00596-8",
"author": "A Chimalakonda",
"doi-asserted-by": "publisher",
"first-page": "1763",
"journal-title": "Dermatol. Ther.",
"key": "506_CR33",
"unstructured": "Chimalakonda, A. et al. Selectivity Profile of the Tyrosine Kinase 2 Inhibitor Deucravacitinib Compared with Janus Kinase 1/2/3 Inhibitors. Dermatol. Ther. 11, 1763–1776 (2021).",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.1080/17512433.2023.2219054",
"author": "S Elyoussfi",
"doi-asserted-by": "publisher",
"first-page": "549",
"journal-title": "Expert Rev. Clin. Pharmacol.",
"key": "506_CR34",
"unstructured": "Elyoussfi, S., Rane, S. S., Eyre, S. & Warren, R. B. TYK2 as a novel therapeutic target in psoriasis. Expert Rev. Clin. Pharmacol. 16, 549–558 (2023).",
"volume": "16",
"year": "2023"
},
{
"author": "B Strobl",
"first-page": "4000",
"journal-title": "J. Immunol. Baltim. Md.",
"key": "506_CR35",
"unstructured": "Strobl, B. et al. Novel functions of tyrosine kinase 2 in the antiviral defense against murine cytomegalovirus. J. Immunol. Baltim. Md. 175, 4000–4008 (2005).",
"volume": "175",
"year": "2005"
},
{
"DOI": "10.1038/s41579-022-00846-2",
"author": "HE Davis",
"doi-asserted-by": "publisher",
"first-page": "133",
"journal-title": "Nat. Rev. Microbiol.",
"key": "506_CR36",
"unstructured": "Davis, H. E., McCorkell, L., Vogel, J. M. & Topol, E. J. Long COVID: major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 21, 133–146 (2023).",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.1371/journal.pgen.1010113",
"author": "A Verma",
"doi-asserted-by": "publisher",
"first-page": "e1010113",
"journal-title": "PLoS Genet",
"key": "506_CR37",
"unstructured": "Verma, A. et al. A Phenome-Wide Association Study of genes associated with COVID-19 severity reveals shared genetics with complex diseases in the Million Veteran Program. PLoS Genet 18, e1010113 (2022).",
"volume": "18",
"year": "2022"
},
{
"DOI": "10.1016/j.ebiom.2021.103277",
"author": "J Fadista",
"doi-asserted-by": "publisher",
"journal-title": "EBioMedicine",
"key": "506_CR38",
"unstructured": "Fadista, J. et al. Shared genetic etiology between idiopathic pulmonary fibrosis and COVID-19 severity. EBioMedicine 65, 103277 (2021).",
"volume": "65",
"year": "2021"
},
{
"DOI": "10.1164/rccm.202109-2166OC",
"author": "A Verma",
"doi-asserted-by": "publisher",
"first-page": "1220",
"journal-title": "Am. J. Respir. Crit. Care Med.",
"key": "506_CR39",
"unstructured": "Verma, A. et al. A MUC5B Gene Polymorphism, rs35705950-T, Confers Protective Effects Against COVID-19 Hospitalization but Not Severe Disease or Mortality. Am. J. Respir. Crit. Care Med. 206, 1220–1229 (2022).",
"volume": "206",
"year": "2022"
},
{
"DOI": "10.5281/ZENODO.10984028",
"doi-asserted-by": "publisher",
"key": "506_CR40",
"unstructured": "pietznerm. comp-med/phecode-covid19-ukb: v1.0. [object Object] https://doi.org/10.5281/ZENODO.10984028 (2024)."
}
],
"reference-count": 40,
"references-count": 40,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.nature.com/articles/s43856-024-00506-x"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Complex patterns of multimorbidity associated with severe COVID-19 and long COVID",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1007/springer_crossmark_policy",
"volume": "4"
}