Cannabidiol inhibits SARS-CoV-2 replication through induction of the host ER stress and innate immune responses
et al., Science Advances, doi:10.1126/sciadv.abi6110, Jan 2022 (preprint)
Retrospective 1,212 patients in the USA with a history of seizure-related conditions, showing patients treated with CBD100 had significantly lower incidence of COVID-19 cases compared to a matched control group.
In vitro study showing CBD inhibits SARS-CoV-2 with Vero E6 and Calu-3 cells. Mouse study showing CBD significantly inhibited viral replication in the lung and nasal turbinate.
Authors note that CBD does not inhibit ACE2 expression or the main viral proteases, inhibition occurs after viral entry. Authors stress several limitations for use at this time, including purity, quality, and the formulation of products, and potential lung damage based on administration method.
Authors recommend clinical trials, but do not mention the existing RCT by Crippa et al.
Standard of Care (SOC) for COVID-19 in the study country,
the USA, is very poor with very low average efficacy for approved treatments1.
Only expensive, high-profit treatments were approved for early treatment. Low-cost treatments were excluded, reducing the probability of early treatment due to access and cost barriers, and eliminating complementary and synergistic benefits seen with many low-cost treatments.
|
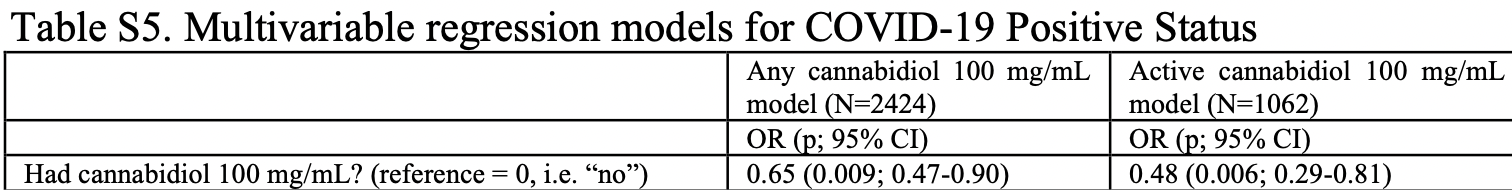
risk of case, 49.6% lower, RR 0.50, p = 0.006, treatment 26 of 531 (4.9%), control 48 of 531 (9.0%), NNT 24, odds ratio converted to relative risk, active CBD100 users.
|
|
risk of case, 32.9% lower, RR 0.67, p = 0.009, treatment 75 of 1,212 (6.2%), control 108 of 1,212 (8.9%), NNT 37, odds ratio converted to relative risk, all CBD100 users.
|
| Effect extraction follows pre-specified rules prioritizing more serious outcomes. Submit updates |
Nguyen et al., 20 Jan 2022, retrospective, USA, peer-reviewed, 34 authors.
Cannabidiol inhibits SARS-CoV-2 replication through induction of the host ER stress and innate immune responses
The spread of SARS-CoV-2 and ongoing COVID-19 pandemic underscores the need for new treatments. Here we report that cannabidiol (CBD) inhibits infection of SARS-CoV-2 in cells and mice. CBD and its metabolite 7-OH-CBD, but not THC or other congeneric cannabinoids tested, potently block SARS-CoV-2 replication in lung epithelial cells. CBD acts after viral entry, inhibiting viral gene expression and reversing many effects of SARS-CoV-2 on host gene transcription. CBD inhibits SARS-CoV-2 replication in part by up-regulating the host IRE1 RNase endoplasmic reticulum (ER) stress response and interferon signaling pathways. In matched groups of human patients from the National COVID Cohort Collaborative, CBD (100 mg/ml oral solution per medical records) had a significant negative association with positive SARS-CoV-2 tests. This study highlights CBD as a potential preventative agent for early-stage SARS-CoV-2 infection and merits future clinical trials. We caution against use of non-medical formulations including edibles, inhalants or topicals as a preventative or treatment therapy at the present time.
SUPPLEMENTARY MATERIALS Supplementary material for this article is available at https://science.org/doi/10.1126/ sciadv.abi6110 View/request a protocol for this paper from Bio-protocol. Teaser: Cannabidiol from the cannabis plant has potential to prevent and inhibit SARS-CoV-2 infection.
of 18 National COVID Cohort Collaborative (N3C). The patient data analyses described in this publication were conducted with data or tools accessed through the NCATS N3C Data Enclave covid.cd2h.org/enclave and supported by NCATS U24 TR002306. This research was possible because of the patients whose information is included within the data from participating organizations (covid.cd2h.org/dtas) and the organizations (https://ncats.nih.gov/n3c/ resources/data-contribution/data-transfer-agreement-signatories) and scientists who have contributed to the on-going development of this community resource (28) . The project described was supported by the National Institute of General Medical Sciences, 5U54GM104942-04. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The analysis used only de-identified data (i.e., N3C Data Access Tier 2, described at https://covid.cd2h.org/N3C_governance). We gratefully acknowledge contributions from the following N3C core teams:
of 18 • Additional N3C acknowledgments. We would also like to thank others not listed above: Joy Alamgir (ARIScience), Seth Russell, MS (University of Colorado),..
References
Administration, None
Afgan, Baker, Batut, Van Den Beek, Bouvier et al., The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update, Nucleic Acids Res
Anil, Shalev, Vinayaka, Nadarajan, Namdar et al., Cannabis compounds exhibit anti-inflammatory activity in vitro in COVID-19-related inflammation in lung epithelial cells and pro-inflammatory activity in macrophages, Sci. Rep
Austin, Balance diagnostics for comparing the distribution of baseline covariates between treatment groups in propensity-score matched samples, Stat. Med
Bechill, Chen, Brewer, Baker, Mouse hepatitis virus infection activates the Ire1/XBP1 pathway of the unfolded protein response, Adv. Exp. Med. Biol
Beltran-Montoya, Herrerias-Canedo, Arzola-Paniagua, Vadillo-Ortega, Dueñas-Garcia et al., A randomized, clinical trial of ketorolac tromethamine vs ketorolac trometamine plus complex B vitamins for cesarean delivery analgesia, Saudi J Anaesth
Blanco-Melo, Nilsson-Payant, Liu, Uhl, Hoagland et al., Imbalanced host response to SARS-CoV-2 drives development of COVID-19, Cell
Blizzard, Hosmer, Parameter estimation and goodness-of-fit in log binomial regression, Biom. J
Center, None
Corp, Tabulate Twoway reference manual
Crawford, Eguia, Dingens, Loes, Malone et al., Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays, Viruses
Daniloski, Jordan, Wessels, Hoagland, Kasela et al., Identification of required host factors for SARS-CoV-2 infection in human cells, Cell
Dash, Ali, Jahan, Munni, Mitra et al., Emerging potential of cannabidiol in reversing proteinopathies, Ageing Res. Rev
Di, Elbahesh, Brinton, Characteristics of human OAS1 isoform proteins, Viruses
Dobin, Davis, Schlesinger, Drenkow, Zaleski et al., STAR: Ultrafast universal RNA-seq aligner, Bioinformatics
Fung, Liu, The ER stress sensor IRE1 and MAP kinase ERK modulate autophagy induction in cells infected with coronavirus infectious bronchitis virus, Virology
Galloway, Paul, Maccannell, Johansson, Brooks et al., Emergence of SARS-CoV-2 B.1.1.7 lineage -United States, MMWR Morb Mortal Wkly Rep
Haendel, Chute, Bennett, Eichmann, Guinney et al., the N3C Consortium, The National COVID Cohort Collaborative (N3C): Rationale, design, infrastructure, and deployment, J. Am. Med. Inform. Assoc
Haller, Kochs, Weber, The interferon response circuit: induction and suppression by pathogenic viruses, Virology
Hepburn, Mullaguri, George, Hantus, Punia et al., Acute symptomatic seizures in critically ill patients with COVID-19: Is there an association? Neurocrit, Care
Hetz, Chevet, Oakes, Proteostasis control by the unfolded protein response, Nat. Cell Biol
Hetz, Martinon, Rodriguez, Glimcher, The unfolded protein response: Integrating stress signals through the stress sensor IRE1, Physiol. Rev
Hetz, Zhang, Kaufman, Mechanisms, regulation and functions of the unfolded protein response, Nat. Rev. Mol. Cell Biol
Ho, Imai, King, Suart, Whitworth et al., MatchIt: Nonparametric Preprocessing for Parametric Causal Inference
Hoffmann, Kleine-Weber, Schroeder, Krüger, Herrler et al., SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor, Cell
Institute, None
Jaimes, Millet, Whittaker, Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 Site, iScience
Kneller, Phillips, O'neill, Jedrzejczak, Stols et al., Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography, Nat. Commun
Kompaniyets, Goodman, Belay, Freedman, Sucosky et al., Body mass index and risk for COVID-19-related hospitalization, intensive care unit admission, invasive mechanical ventilation, and Death -United States, MMWR Morb. Mortal. Wkly Rep
Krueger, Galore, A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files
Leas, Hendrickson, Nobles, Todd, Smith et al., Self-reported Cannabidiol (CBD) use for conditions with proven therapies, JAMA Netw. Open
Liao, Smyth, Shi, featureCounts: An efficient general purpose program for assigning sequence reads to genomic features, Bioinformatics
Love, Huber, Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol
Lowe, Toyang, Mclaughlin, Potential of cannabidiol for the treatment of viral hepatitis, Pharm. Res
Matsuyama, Nao, Shirato, Kawase, Saito et al., Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells, Proc. Natl. Acad. Sci. U.S.A
Mccray, Pewe, Wohlford-Lenane, Hickey, Manzel et al., Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus, J. Virol
Morimoto, Hay, Sperling, Chen, Lee et al., We thank the University of Chicago Genomics Facility (RRID:SCR_019196) especially Sandhiya Arun and Pieter Faber, for their assistance with RNA sequencing. Finally, we would like to acknowledge the University of Chicago Vice Provost for Research
Muthumalage, Rahman, Cannabidiol differentially regulates basal and LPS-induced inflammatory responses in macrophages, lung epithelial cells, and fibroblasts, Toxicol. Appl. Pharmacol
Nelson, Bisson, Singh, Graham, Chen et al., The essential medicinal chemistry of cannabidiol (CBD), J. Med. Chem
Nguyen, Teaser: Cannabidiol from the cannabis plant has potential to prevent and inhibit SARS-CoV-2 infection, IMMUNOLOGY, LIFE SCIENCES Emp
Osipiuk, Azizi, Dvorkin, Endres, Jedrzejczak et al., Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors, Nat. Commun
Pregibon, Data Analytic Methods for Generalized Linear Models
Reed, Muench, A simple method of estimating fifty per cent endpoints, Am. J. Epidemiol
Robinson, Thorvaldsdóttir, Winckler, Guttman, Lander et al., Integrative genomics viewer, Nat. Biotechnol
Schneider, Luna, Hoffmann, Sánchez-Rivera, Leal et al., Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks, Cell
Sekar, Pack, Epidiolex as adjunct therapy for treatment of refractory epilepsy: A comprehensive review with a focus on adverse effects, F1000Res
Shahbazi, Grandi, Banerjee, Trant, Cannabinoids and cannabinoid receptors: The story so far, iScience
Sorkin, Kuszak, Bloss, Fukagawa, Hoffman et al., Improving natural product research translation: From source to clinical trial, FASEB J
Subramanian, Tamayo, Mootha, Mukherjee, Ebert et al., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles, Proc. Natl. Acad. Sci. U.S.A
Taylor, Gidal, Blakey, Tayo, Morrison, A phase I, randomized, double-blind, placebo-controlled, single ascending dose, multiple dose, and food effect trial of the safety, tolerability and pharmacokinetics of highly purified cannabidiol in healthy subjects, CNS Drugs
Tummino, Rezelj, Fischer, Fischer, O'meara et al., Phospholipidosis is a shared mechanism underlying the in vitro antiviral activity of many repurposed drugs against SARS-CoV-2, bioRxiv
Wang, Simoneau, Kulsuptrakul, Bouhaddou, Travisano et al., Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses, Cell
Weir, Jan, StatPearls
Weiss, Leibowitz, Coronavirus pathogenesis, Adv. Virus Res
Yoon, Park, Choi, Yang, Jeong et al., Real-time PCR quantification of spliced X-box binding protein 1 (XBP1) using a universal primer method, PLOS ONE
Zhou, Chen, Shannon, Wei, Xiang et al., Interferon-2b Treatment for COVID-19, Front. Immunol
Zhou, Zhou, Pache, Chang, Khodabakhshi et al., Metascape provides a biologist-oriented resource for the analysis of systems-level datasets, Nat. Commun
DOI record:
{
"DOI": "10.1126/sciadv.abi6110",
"ISSN": [
"2375-2548"
],
"URL": "http://dx.doi.org/10.1126/sciadv.abi6110",
"abstract": "<jats:p>The spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and ongoing coronavirus disease 2019 (COVID-19) pandemic underscores the need for new treatments. Here, we report that cannabidiol (CBD) inhibits infection of SARS-CoV-2 in cells and mice. CBD and its metabolite 7-OH-CBD, but not THC or other congeneric cannabinoids tested, potently block SARS-CoV-2 replication in lung epithelial cells. CBD acts after viral entry, inhibiting viral gene expression and reversing many effects of SARS-CoV-2 on host gene transcription. CBD inhibits SARS-CoV-2 replication in part by up-regulating the host IRE1α ribonuclease endoplasmic reticulum (ER) stress response and interferon signaling pathways. In matched groups of human patients from the National COVID Cohort Collaborative, CBD (100 mg/ml oral solution per medical records) had a significant negative association with positive SARS-CoV-2 tests. This study highlights CBD as a potential preventative agent for early-stage SARS-CoV-2 infection and merits future clinical trials. We caution against current use of non-medical formulations as a preventative or treatment therapy.</jats:p>",
"alternative-id": [
"10.1126/sciadv.abi6110"
],
"author": [
{
"ORCID": "http://orcid.org/0000-0002-4637-2331",
"affiliation": [
{
"name": "Ben May Department for Cancer Research, University of Chicago, Chicago, IL 60637, USA."
}
],
"authenticated-orcid": true,
"family": "Nguyen",
"given": "Long Chi",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0002-8343-5477",
"affiliation": [
{
"name": "Ben May Department for Cancer Research, University of Chicago, Chicago, IL 60637, USA."
}
],
"authenticated-orcid": true,
"family": "Yang",
"given": "Dongbo",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-6347-0232",
"affiliation": [
{
"name": "Department of Microbiology, University of Chicago, Chicago, IL 60637, USA."
},
{
"name": "Howard Taylor Ricketts Laboratory, Argonne National Laboratory, Lemont, IL 60439, USA."
}
],
"authenticated-orcid": true,
"family": "Nicolaescu",
"given": "Vlad",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-0654-6250",
"affiliation": [
{
"name": "Center for Health and the Social Sciences, University of Chicago, Chicago, IL 60637, USA."
}
],
"authenticated-orcid": true,
"family": "Best",
"given": "Thomas J.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-8475-7435",
"affiliation": [
{
"name": "Department of Microbiology, University of Chicago, Chicago, IL 60637, USA."
},
{
"name": "Howard Taylor Ricketts Laboratory, Argonne National Laboratory, Lemont, IL 60439, USA."
}
],
"authenticated-orcid": true,
"family": "Gula",
"given": "Haley",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Center for Predictive Medicine for Biodefense and Emerging Infectious Diseases, University of Louisville, Louisville, KY 40222, USA."
}
],
"family": "Saxena",
"given": "Divyasha",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Center for Predictive Medicine for Biodefense and Emerging Infectious Diseases, University of Louisville, Louisville, KY 40222, USA."
}
],
"family": "Gabbard",
"given": "Jon D.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-0748-0863",
"affiliation": [
{
"name": "Pharmacognosy Institute and Department of Pharmaceutical Sciences, College of Pharmacy, University of Illinois at Chicago, Chicago, IL 60612, USA."
}
],
"authenticated-orcid": true,
"family": "Chen",
"given": "Shao-Nong",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Pharmacognosy Institute and Department of Pharmaceutical Sciences, College of Pharmacy, University of Illinois at Chicago, Chicago, IL 60612, USA."
}
],
"family": "Ohtsuki",
"given": "Takashi",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-0739-9224",
"affiliation": [
{
"name": "Pharmacognosy Institute and Department of Pharmaceutical Sciences, College of Pharmacy, University of Illinois at Chicago, Chicago, IL 60612, USA."
}
],
"authenticated-orcid": true,
"family": "Friesen",
"given": "John Brent",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-4460-9558",
"affiliation": [
{
"name": "Pritzker School of Molecular Engineering, University of Chicago, Chicago, IL 60637, USA."
}
],
"authenticated-orcid": true,
"family": "Drayman",
"given": "Nir",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Pritzker School of Molecular Engineering, University of Chicago, Chicago, IL 60637, USA."
}
],
"family": "Mohamed",
"given": "Adil",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Ben May Department for Cancer Research, University of Chicago, Chicago, IL 60637, USA."
}
],
"family": "Dann",
"given": "Christopher",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pathology, University of Chicago, Chicago, IL 60637, USA."
}
],
"family": "Silva",
"given": "Diane",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Ben May Department for Cancer Research, University of Chicago, Chicago, IL 60637, USA."
}
],
"family": "Robinson-Mailman",
"given": "Lydia",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Ben May Department for Cancer Research, University of Chicago, Chicago, IL 60637, USA."
}
],
"family": "Valdespino",
"given": "Andrea",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-6302-1373",
"affiliation": [
{
"name": "Ben May Department for Cancer Research, University of Chicago, Chicago, IL 60637, USA."
}
],
"authenticated-orcid": true,
"family": "Stock",
"given": "Letícia",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Ben May Department for Cancer Research, University of Chicago, Chicago, IL 60637, USA."
}
],
"family": "Suárez",
"given": "Eva",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-0711-5516",
"affiliation": [
{
"name": "Department of Chemistry, University of Chicago, Chicago, IL 60637, USA."
}
],
"authenticated-orcid": true,
"family": "Jones",
"given": "Krysten A.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-9226-9917",
"affiliation": [
{
"name": "Department of Chemistry, University of Chicago, Chicago, IL 60637, USA."
}
],
"authenticated-orcid": true,
"family": "Azizi",
"given": "Saara-Anne",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-4807-6250",
"affiliation": [
{
"name": "Center for Predictive Medicine for Biodefense and Emerging Infectious Diseases, University of Louisville, Louisville, KY 40222, USA."
}
],
"authenticated-orcid": true,
"family": "Demarco",
"given": "Jennifer K.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Center for Predictive Medicine for Biodefense and Emerging Infectious Diseases, University of Louisville, Louisville, KY 40222, USA."
}
],
"family": "Severson",
"given": "William E.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Center for Predictive Medicine for Biodefense and Emerging Infectious Diseases, University of Louisville, Louisville, KY 40222, USA."
}
],
"family": "Anderson",
"given": "Charles D.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5361-5776",
"affiliation": [
{
"name": "Department of Surgery, University of Chicago, Chicago, IL 60637, USA."
}
],
"authenticated-orcid": true,
"family": "Millis",
"given": "James Michael",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-9616-1911",
"affiliation": [
{
"name": "Department of Chemistry, University of Chicago, Chicago, IL 60637, USA."
}
],
"authenticated-orcid": true,
"family": "Dickinson",
"given": "Bryan C.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-1912-6020",
"affiliation": [
{
"name": "Pritzker School of Molecular Engineering, University of Chicago, Chicago, IL 60637, USA."
}
],
"authenticated-orcid": true,
"family": "Tay",
"given": "Savaş",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-2099-2249",
"affiliation": [
{
"name": "Department of Pathology, University of Chicago, Chicago, IL 60637, USA."
}
],
"authenticated-orcid": true,
"family": "Oakes",
"given": "Scott A.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-1022-4326",
"affiliation": [
{
"name": "Pharmacognosy Institute and Department of Pharmaceutical Sciences, College of Pharmacy, University of Illinois at Chicago, Chicago, IL 60612, USA."
}
],
"authenticated-orcid": true,
"family": "Pauli",
"given": "Guido F.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-2811-1111",
"affiliation": [
{
"name": "Center for Predictive Medicine for Biodefense and Emerging Infectious Diseases, University of Louisville, Louisville, KY 40222, USA."
}
],
"authenticated-orcid": true,
"family": "Palmer",
"given": "Kenneth E.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-2790-7393",
"affiliation": [
{
"name": "Center for Health and the Social Sciences, University of Chicago, Chicago, IL 60637, USA."
}
],
"authenticated-orcid": true,
"family": "Meltzer",
"given": "David O.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-7417-3615",
"affiliation": [
{
"name": "Department of Microbiology, University of Chicago, Chicago, IL 60637, USA."
},
{
"name": "Howard Taylor Ricketts Laboratory, Argonne National Laboratory, Lemont, IL 60439, USA."
}
],
"authenticated-orcid": true,
"family": "Randall",
"given": "Glenn",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-6586-8335",
"affiliation": [
{
"name": "Ben May Department for Cancer Research, University of Chicago, Chicago, IL 60637, USA."
}
],
"authenticated-orcid": true,
"family": "Rosner",
"given": "Marsha Rich",
"sequence": "additional"
},
{
"affiliation": [],
"name": "The National COVID Cohort Collaborative Consortium",
"sequence": "additional"
}
],
"container-title": "Science Advances",
"container-title-short": "Sci. Adv.",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2022,
2,
23
]
],
"date-time": "2022-02-23T18:59:33Z",
"timestamp": 1645642773000
},
"deposited": {
"date-parts": [
[
2024,
1,
9
]
],
"date-time": "2024-01-09T19:37:26Z",
"timestamp": 1704829046000
},
"indexed": {
"date-parts": [
[
2024,
4,
6
]
],
"date-time": "2024-04-06T09:02:08Z",
"timestamp": 1712394128544
},
"is-referenced-by-count": 68,
"issue": "8",
"issued": {
"date-parts": [
[
2022,
2,
25
]
]
},
"journal-issue": {
"issue": "8",
"published-print": {
"date-parts": [
[
2022,
2,
25
]
]
}
},
"language": "en",
"link": [
{
"URL": "https://www.science.org/doi/pdf/10.1126/sciadv.abi6110",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "221",
"original-title": [],
"prefix": "10.1126",
"published": {
"date-parts": [
[
2022,
2,
25
]
]
},
"published-print": {
"date-parts": [
[
2022,
2,
25
]
]
},
"publisher": "American Association for the Advancement of Science (AAAS)",
"reference": [
{
"DOI": "10.1016/B978-0-12-385885-6.00009-2",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_2_2"
},
{
"DOI": "10.15585/mmwr.mm7003e2",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_3_2"
},
{
"DOI": "10.1016/j.cell.2020.02.052",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_4_2"
},
{
"DOI": "10.1016/j.isci.2020.101212",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_5_2"
},
{
"DOI": "10.1073/pnas.2002589117",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_6_2"
},
{
"DOI": "10.1016/j.cell.2020.12.004",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_7_2"
},
{
"DOI": "10.1016/j.cell.2020.12.006",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_8_2"
},
{
"DOI": "10.1016/j.cell.2020.10.030",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_9_2"
},
{
"DOI": "10.1038/s41467-020-16954-7",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_10_2"
},
{
"DOI": "10.1038/s41467-021-21060-3",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_11_2"
},
{
"DOI": "10.1016/j.isci.2020.101301",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_12_2"
},
{
"DOI": "10.1021/acs.jmedchem.0c00724",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_13_2"
},
{
"DOI": "10.12688/f1000research.16515.1",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_14_2"
},
{
"article-title": "Potential of cannabidiol for the treatment of viral hepatitis",
"author": "Lowe H. I.",
"first-page": "116",
"journal-title": "Pharm. Res.",
"key": "e_1_3_3_15_2",
"unstructured": "H. I. Lowe, N. J. Toyang, W. McLaughlin, Potential of cannabidiol for the treatment of viral hepatitis. Pharm. Res. 9, 116–118 (2017).",
"volume": "9",
"year": "2017"
},
{
"DOI": "10.1007/s40263-018-0578-5",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_16_2"
},
{
"DOI": "10.1038/s41598-021-81049-2",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_17_2"
},
{
"DOI": "10.3390/v12050513",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_18_2"
},
{
"DOI": "10.1038/ncb3184",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_19_2"
},
{
"DOI": "10.1016/j.arr.2020.101209",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_20_2"
},
{
"DOI": "10.1007/978-0-387-33012-9_24",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_21_2"
},
{
"DOI": "10.1016/j.virol.2019.05.002",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_22_2"
},
{
"DOI": "10.1038/s41580-020-0250-z",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_23_2"
},
{
"DOI": "10.1016/j.virol.2005.09.024",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_24_2"
},
{
"DOI": "10.1016/j.cell.2020.04.026",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_25_2"
},
{
"DOI": "10.3390/v12020152",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_26_2"
},
{
"article-title": "Phospholipidosis is a shared mechanism underlying the in vitro antiviral activity of many repurposed drugs against SARS-CoV-2",
"author": "Tummino T. A.",
"journal-title": "bioRxiv",
"key": "e_1_3_3_27_2",
"unstructured": "T. A. Tummino, V. V. Rezelj, B. Fischer, A. Fischer, M. J. O’Meara, B. Monel, T. Vallet, Z. Zhang, A. Alon, H. R. O’Donnell, J. Lyu, H. Schadt, K. M. White, N. J. Krogan, L. Urban, K. M. Shokat, A. C. Kruse, A. García-Sastre, O. Schwartz, F. Moretti, M. Vignuzzi, F. Pognan, B. K. Shoichet, Phospholipidosis is a shared mechanism underlying the in vitro antiviral activity of many repurposed drugs against SARS-CoV-2. bioRxiv 10.1101/2021.03.23.436648 , (2021).",
"year": "2021"
},
{
"DOI": "10.1128/JVI.02012-06",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_28_2"
},
{
"DOI": "10.1093/jamia/ocaa196",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_29_2"
},
{
"key": "e_1_3_3_30_2",
"unstructured": "Centers for Disease Control and Prevention COVID-19 People at Increased Risk People with Certain Medical Conditions (CDC 2021)."
},
{
"DOI": "10.3389/fimmu.2020.01061",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_31_2"
},
{
"DOI": "10.1016/j.taap.2019.114713",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_32_2"
},
{
"key": "e_1_3_3_33_2",
"unstructured": "Food and Drug Administration (FDA) Application number 210365Orig1s000 Clinical Pharmacology and Biopharmaceutics Reviews (2017)."
},
{
"DOI": "10.1096/fj.201902143R",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_34_2"
},
{
"DOI": "10.1093/nar/gky379",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_35_2"
},
{
"key": "e_1_3_3_36_2",
"unstructured": "F. Krueger Trim Galore A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. (2015); https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/."
},
{
"DOI": "10.1093/bioinformatics/bts635",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_37_2"
},
{
"DOI": "10.1093/bioinformatics/btt656",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_38_2"
},
{
"DOI": "10.1186/s13059-014-0550-8",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_39_2"
},
{
"DOI": "10.1038/nbt.1754",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_40_2"
},
{
"DOI": "10.1371/journal.pone.0219978",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_41_2"
},
{
"DOI": "10.1038/s41467-019-09234-6",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_42_2"
},
{
"DOI": "10.1073/pnas.0506580102",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_43_2"
},
{
"DOI": "10.1152/physrev.00001.2011",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_44_2"
},
{
"DOI": "10.1093/oxfordjournals.aje.a118408",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_45_2"
},
{
"key": "e_1_3_3_46_2",
"unstructured": "Observational Health Data Sciences and Informatics ATLAS (2021); www.ohdsi.org/software-tools/atlas/."
},
{
"key": "e_1_3_3_47_2",
"unstructured": "Observational Medical Outcomes Partnership (OMOP) OMOP Clinical Data Model v5.3.1 Standardized Derived Elements Drug Era table (2021); https://ohdsi.github.io/CommonDataModel/cdm53.html#Standardized_Derived_Elements."
},
{
"key": "e_1_3_3_48_2",
"unstructured": "Centers for Disease Control and Prevention COVID-19: When to Quarantine (CDC 2021)."
},
{
"key": "e_1_3_3_49_2",
"unstructured": "Text Processing Services Library Regular Expression Operations (Python Software Foundation 2021); https://docs.python.org/3/library/re.html."
},
{
"DOI": "10.4103/1658-354X.101209",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_50_2"
},
{
"key": "e_1_3_3_51_2",
"unstructured": "Centers for Disease Control and Prevention COVID-19-Hospitalization and Death by Age (CDC 2021)."
},
{
"key": "e_1_3_3_52_2",
"unstructured": "Observational Medical Outcomes Partnership (OMOP) OMOP Clinical Data Model v5.3.1 Clinical Data Tables Person table (2021); https://ohdsi.github.io/CommonDataModel/cdm53.html."
},
{
"DOI": "10.15585/mmwr.mm7010e4",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_53_2"
},
{
"key": "e_1_3_3_54_2",
"unstructured": "C. B. Weir A. Jan BMI classification percentile and cut off points. In: StatPearls [ Internet ] (StatPearls Publishing January 2021 updated 9 May 2021); https://www.ncbi.nlm.nih.gov/books/NBK541070/."
},
{
"key": "e_1_3_3_55_2",
"unstructured": "US Food and Drug Administration FDA approves new indication for drug containing an active ingredient derived from cannabis to treat seizures in rare genetic Disease (FDA news release 31 July 2020); https://www.fda.gov/news-events/press-announcements/fda-approves-new-indication-drug-containing-active-ingredient-derived-cannabis-treat-seizures-rare."
},
{
"DOI": "10.1001/jamanetworkopen.2020.20977",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_56_2"
},
{
"DOI": "10.1007/s12028-020-01006-1",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_57_2"
},
{
"key": "e_1_3_3_58_2",
"unstructured": "StataCorp Tabulate two-way reference manual (2021); https://www.stata.com/manuals/rtabulatetwoway.pdf."
},
{
"article-title": "MatchIt: nonparametric preprocessing for parametric causal inference",
"author": "Ho D.",
"first-page": "1",
"journal-title": "J. Stat. Softw.",
"key": "e_1_3_3_59_2",
"unstructured": "D. Ho, K. Imai, G. King, E. A. Stuart, MatchIt: nonparametric preprocessing for parametric causal inference. J. Stat. Softw. 42, 1–28 (2021).",
"volume": "42",
"year": "2021"
},
{
"DOI": "10.1002/sim.3697",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_60_2"
},
{
"key": "e_1_3_3_61_2",
"unstructured": "D. Pregibon Data Analytic Methods for Generalized Linear Models. Thesis University of Toronto (1979)."
},
{
"DOI": "10.1002/bimj.200410165",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_62_2"
},
{
"key": "e_1_3_3_63_2",
"unstructured": "John Hopkins Coronavirus Resource Center Covid-19 Dashboard. (2021); https://coronavirus.jhu.edu/map.html."
},
{
"key": "e_1_3_3_64_2",
"unstructured": "National COVID Cohort Collaborative COVID-19 Phenotype 3.0 Additional Information (N3C 2021); https://github.com/National-COVID-Cohort-Collaborative/Phenotype_Data_Acquisition/wiki/Phenotype-3.0."
}
],
"reference-count": 63,
"references-count": 63,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.science.org/doi/10.1126/sciadv.abi6110"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Multidisciplinary"
],
"subtitle": [],
"title": "Cannabidiol inhibits SARS-CoV-2 replication through induction of the host ER stress and innate immune responses",
"type": "journal-article",
"volume": "8"
}