A machine learning-based phenotype for long COVID in children: an EHR-based study from the RECOVER program
et al., medRxiv, doi:10.1101/2022.12.22.22283791, Dec 2022
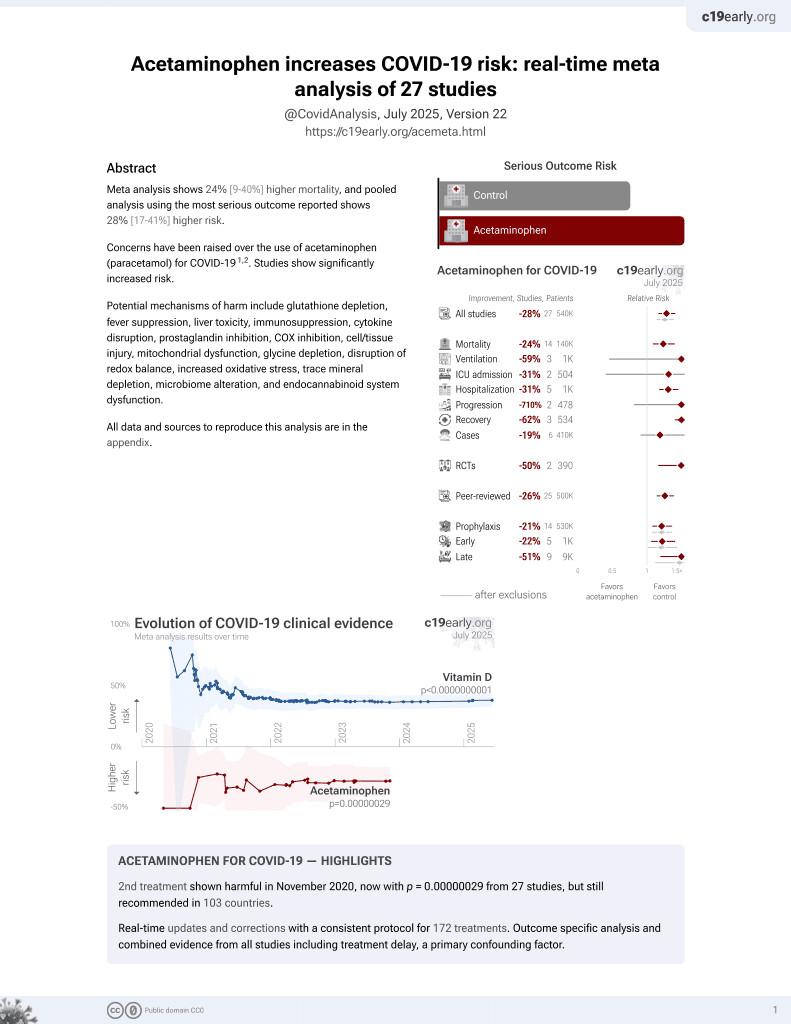
2nd treatment shown to increase risk in
November 2020, now with p = 0.00000029 from 27 studies, but still recommended in 103 countries.
6,400+ studies for
210+ treatments. c19early.org
|
Retrospective 87,398 pediatric patients in the USA, reporting acetaminophen and aspirin associated with PASC, without specific details. Authors note that this could be related to use for MIS-C treatment.
Acetaminophen is also known as paracetamol, Tylenol, Panadol, Calpol, Tempra, Calprofen, Doliprane, Efferalgan, Grippostad C, Dolo, Acamol, Fevadol, Crocin, and Perfalgan.
Lorman et al., 26 Dec 2022, retrospective, USA, preprint, 18 authors, study period September 2021 - April 2022.
A machine learning-based phenotype for long COVID in children: an EHR-based study from the RECOVER program Authors
doi:10.1101/2022.12.22.22283791
Background As clinical understanding of pediatric Post-Acute Sequelae of SARS CoV-2 (PASC) develops, and hence the clinical definition evolves, it is desirable to have a method to reliably identify patients who are likely to have post-acute sequelae of SARS CoV-2 (PASC) in health systems data.
Methods and Findings In this study, we developed and validated a machine learning algorithm to classify which patients have PASC (distinguishing between Multisystem Inflammatory Syndrome in Children (MIS-C) and non-MIS-C variants) from a cohort of patients with positive SARS-CoV-2 test results in pediatric health systems within the PEDSnet EHR network. Patient features included in the model were selected from conditions, procedures, performance of diagnostic testing, and medications using a tree-based scan statistic approach. We used an XGboost model, with hyperparameters selected through cross-validated grid search, and model performance was assessed using 5-fold cross-validation. Model predictions and feature importance were evaluated using Shapley Additive exPlanation (SHAP) values.
Conclusions The model provides a tool for identifying patients with PASC and an approach to characterizing PASC using diagnosis, medication, laboratory, and procedure features in health systems data. Using appropriate threshold settings, the model can be used to identify PASC patients in health systems data at higher precision for inclusion in studies or at higher recall in screening for clinical trials, especially in settings where PASC diagnosis codes are used less frequently or less reliably. Analysis of how specific features contribute to the classification process may assist in gaining a better understanding of features that are associated with PASC diagnoses.
References
Algarni, Alamri, Khayat, Alabdali, Alsubhi et al., Clinical practice guidelines in multisystem inflammatory syndrome (MIS-C) related to COVID-19: a critical review and recommendations, World J Pediatr, doi:10.1007/s12519-021-00499-w
Borch, Holm, Knudsen, Ellermann-Eriksen, Hagstroem, Long COVID symptoms and duration in SARS-CoV-2 positive children -a nationwide cohort study, Eur J Pediatr, doi:10.1007/s00431-021-04345-z
Fainardi, Meoli, Chiopris, Long COVID in Children and Adolescents, Life Basel Switz, doi:10.3390/life12020285
Kulldorff, Fang, Walsh, A tree-based scan statistic for database disease surveillance, Biometrics, doi:10.1111/1541-0420.00039
Lundberg, Lee, A Unified Approach to Interpreting Model Predictions
Mahmoud, El-Kalliny, Kotby, El-Ganzoury, Fouda et al., Treatment of MIS-C in Children and Adolescents, Curr Pediatr Rep, doi:10.1007/s40124-021-00259-4
Pellegrino, Chiappini, Licari, Galli, Marseglia, Prevalence and clinical presentation of long COVID in children: a systematic review, Eur J Pediatr, doi:10.1007/s00431-022-04600-x
Pfaff, Girvin, Bennett, Identifying who has long COVID in the USA: a machine learning approach using N3C data, Lancet Digit Health, doi:10.1016/S2589-7500(22)00048-6
Ramakrishnan, Kashour, Hamid, Halwani, Tleyjeh, Unraveling the Mystery Surrounding Post-Acute Sequelae of COVID-19, Front Immunol, doi:10.3389/fimmu.2021.686029
Rao, Lee, Razzaghi, Clinical features and burden of post-acute sequelae of SARS-CoV-2 infection in children and adolescents: an exploratory EHR-based cohort study from the RECOVER program, MedRxiv Prepr Serv Health Sci. Published online, doi:10.1101/2022.05.24.22275544
Reese, Blau, Bergquist, Generalizable Long COVID Subtypes: Findings from the NIH N3C and RECOVER Programs, MedRxiv Prepr Serv Health Sci. Published online, doi:10.1101/2022.05.24.22275398
Saito, Rehmsmeier, The Precision-Recall Plot Is More Informative than the ROC Plot When Evaluating Binary Classifiers on Imbalanced Datasets, PLoS ONE, doi:10.1371/journal.pone.0118432
Thallapureddy, Thallapureddy, Zerda, Long-Term Complications of COVID-19 Infection in Adolescents and Children, Curr Pediatr Rep, doi:10.1007/s40124-021-00260-x
Wang, Maro, Baro, Data Mining for Adverse Drug Events With a Propensity Scorematched Tree-based Scan Statistic, Epidemiol Camb Mass, doi:10.1097/EDE.0000000000000907
Yang, Varghese, Stephenson, Tu, Gronsbell, Machine learning approaches for electronic health records phenotyping: A methodical review, Published online, doi:10.1101/2022.04.23.22274218
DOI record:
{
"DOI": "10.1101/2022.12.22.22283791",
"URL": "http://dx.doi.org/10.1101/2022.12.22.22283791",
"abstract": "<jats:title>Abstract</jats:title><jats:sec><jats:title>Background</jats:title><jats:p>As clinical understanding of pediatric Post-Acute Sequelae of SARS CoV-2 (PASC) develops, and hence the clinical definition evolves, it is desirable to have a method to reliably identify patients who are likely to have post-acute sequelae of SARS CoV-2 (PASC) in health systems data.</jats:p></jats:sec><jats:sec><jats:title>Methods and Findings</jats:title><jats:p>In this study, we developed and validated a machine learning algorithm to classify which patients have PASC (distinguishing between Multisystem Inflammatory Syndrome in Children (MIS-C) and non-MIS-C variants) from a cohort of patients with positive SARS-CoV-2 test results in pediatric health systems within the PEDSnet EHR network. Patient features included in the model were selected from conditions, procedures, performance of diagnostic testing, and medications using a tree-based scan statistic approach. We used an XGboost model, with hyperparameters selected through cross-validated grid search, and model performance was assessed using 5-fold cross-validation. Model predictions and feature importance were evaluated using Shapley Additive exPlanation (SHAP) values.</jats:p></jats:sec><jats:sec><jats:title>Conclusions</jats:title><jats:p>The model provides a tool for identifying patients with PASC and an approach to characterizing PASC using diagnosis, medication, laboratory, and procedure features in health systems data. Using appropriate threshold settings, the model can be used to identify PASC patients in health systems data at higher precision for inclusion in studies or at higher recall in screening for clinical trials, especially in settings where PASC diagnosis codes are used less frequently or less reliably. Analysis of how specific features contribute to the classification process may assist in gaining a better understanding of features that are associated with PASC diagnoses.</jats:p></jats:sec><jats:sec><jats:title>Funding Source</jats:title><jats:p>This research was funded by the National Institutes of Health (NIH) Agreement OT2HL161847-01 as part of the Researching COVID to Enhance Recovery (RECOVER) program of research.</jats:p></jats:sec><jats:sec><jats:title>Disclaimer</jats:title><jats:p>The content is solely the responsibility of the authors and does not necessarily represent the official views of the RECOVER Program, the NIH or other funders.</jats:p></jats:sec>",
"accepted": {
"date-parts": [
[
2022,
12,
26
]
]
},
"author": [
{
"ORCID": "http://orcid.org/0000-0003-0561-6587",
"affiliation": [],
"authenticated-orcid": false,
"family": "Lorman",
"given": "Vitaly",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0001-5001-1590",
"affiliation": [],
"authenticated-orcid": false,
"family": "Razzaghi",
"given": "Hanieh",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Song",
"given": "Xing",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-8307-1642",
"affiliation": [],
"authenticated-orcid": false,
"family": "Morse",
"given": "Keith",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-2189-108X",
"affiliation": [],
"authenticated-orcid": false,
"family": "Utidjian",
"given": "Levon",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-9846-0280",
"affiliation": [],
"authenticated-orcid": false,
"family": "Allen",
"given": "Andrea J.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-0334-6301",
"affiliation": [],
"authenticated-orcid": false,
"family": "Rao",
"given": "Suchitra",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-5251-2399",
"affiliation": [],
"authenticated-orcid": false,
"family": "Rogerson",
"given": "Colin",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-1483-4236",
"affiliation": [],
"authenticated-orcid": false,
"family": "Bennett",
"given": "Tellen D.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-9678-5564",
"affiliation": [],
"authenticated-orcid": false,
"family": "Morizono",
"given": "Hiroki",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Eckrich",
"given": "Daniel",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-9921-0419",
"affiliation": [],
"authenticated-orcid": false,
"family": "Jhaveri",
"given": "Ravi",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-3265-9902",
"affiliation": [],
"authenticated-orcid": false,
"family": "Huang",
"given": "Yungui",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ranade",
"given": "Daksha",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-1637-4942",
"affiliation": [],
"authenticated-orcid": false,
"family": "Pajor",
"given": "Nathan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Lee",
"given": "Grace M.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-1252-068X",
"affiliation": [],
"authenticated-orcid": false,
"family": "Forrest",
"given": "Christopher B.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-8967-0662",
"affiliation": [],
"authenticated-orcid": false,
"family": "Bailey",
"given": "L. Charles",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2022,
12,
26
]
],
"date-time": "2022-12-26T23:30:13Z",
"timestamp": 1672097413000
},
"deposited": {
"date-parts": [
[
2022,
12,
28
]
],
"date-time": "2022-12-28T10:00:33Z",
"timestamp": 1672221633000
},
"group-title": "Epidemiology",
"indexed": {
"date-parts": [
[
2022,
12,
29
]
],
"date-time": "2022-12-29T05:55:56Z",
"timestamp": 1672293356631
},
"institution": [
{
"name": "medRxiv"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2022,
12,
26
]
]
},
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2022.12.22.22283791",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2022,
12,
26
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2022,
12,
26
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference": [
{
"DOI": "10.3390/life12020285",
"doi-asserted-by": "publisher",
"key": "2022122802000890000_2022.12.22.22283791v1.1"
},
{
"DOI": "10.1007/s40124-021-00260-x",
"doi-asserted-by": "publisher",
"key": "2022122802000890000_2022.12.22.22283791v1.2"
},
{
"DOI": "10.1101/2022.05.24.22275544",
"doi-asserted-by": "publisher",
"key": "2022122802000890000_2022.12.22.22283791v1.3"
},
{
"DOI": "10.1101/2022.05.24.22275398",
"doi-asserted-by": "publisher",
"key": "2022122802000890000_2022.12.22.22283791v1.4"
},
{
"DOI": "10.1016/S2589-7500(22)00048-6",
"doi-asserted-by": "publisher",
"key": "2022122802000890000_2022.12.22.22283791v1.5"
},
{
"DOI": "10.1101/2022.04.23.22274218",
"doi-asserted-by": "publisher",
"key": "2022122802000890000_2022.12.22.22283791v1.6"
},
{
"key": "2022122802000890000_2022.12.22.22283791v1.7",
"unstructured": "HAN Archive - 00432 | Health Alert Network (HAN). Published September 21, 2021. Accessed August 18, 2022. https://emergency.cdc.gov/han/2020/han00432.asp"
},
{
"DOI": "10.1007/s12519-021-00499-w",
"doi-asserted-by": "publisher",
"key": "2022122802000890000_2022.12.22.22283791v1.8"
},
{
"DOI": "10.1007/s40124-021-00259-4",
"doi-asserted-by": "publisher",
"key": "2022122802000890000_2022.12.22.22283791v1.9"
},
{
"DOI": "10.1007/s00431-021-04345-z",
"doi-asserted-by": "publisher",
"key": "2022122802000890000_2022.12.22.22283791v1.10"
},
{
"DOI": "10.3389/fimmu.2021.686029",
"doi-asserted-by": "publisher",
"key": "2022122802000890000_2022.12.22.22283791v1.11"
},
{
"DOI": "10.1111/1541-0420.00039",
"doi-asserted-by": "publisher",
"key": "2022122802000890000_2022.12.22.22283791v1.12"
},
{
"key": "2022122802000890000_2022.12.22.22283791v1.13",
"unstructured": "CDC. Healthcare Workers. Centers for Disease Control and Prevention. Published February 11, 2020. Accessed August 18, 2022. https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-care/post-covid-public-health-recs.html"
},
{
"key": "2022122802000890000_2022.12.22.22283791v1.14",
"unstructured": "Coding Long COVID: Characterizing a new disease through an ICD-10 lens | medRxiv. Accessed August 18, 2022. https://www.medrxiv.org/content/10.1101/2022.04.18.22273968v1"
},
{
"key": "2022122802000890000_2022.12.22.22283791v1.15",
"unstructured": "CDC Announces Approval of ICD-10 Code for Post-Acute Sequelae of COVID-19. aapmr.org. Accessed August 22, 2022. https://www.aapmr.org/members-publications/member-news/member-news-details/2021/07/20/cdc-announces-approval-of-icd-10-code-for-post-acute-sequelae-of-covid-19"
},
{
"DOI": "10.1097/EDE.0000000000000907",
"doi-asserted-by": "publisher",
"key": "2022122802000890000_2022.12.22.22283791v1.16"
},
{
"DOI": "10.1371/journal.pone.0118432",
"doi-asserted-by": "publisher",
"key": "2022122802000890000_2022.12.22.22283791v1.17"
},
{
"key": "2022122802000890000_2022.12.22.22283791v1.18",
"unstructured": "Lundberg SM , Lee SI. A Unified Approach to Interpreting Model Predictions. In: Advances in Neural Information Processing Systems. Vol 30. Curran Associates, Inc.; 2017. Accessed August 18, 2022. https://proceedings.neurips.cc/paper/2017/hash/8a20a8621978632d76c43dfd28b67767-Abstract.html"
},
{
"DOI": "10.7717/peerj-cs.880",
"doi-asserted-by": "crossref",
"key": "2022122802000890000_2022.12.22.22283791v1.19",
"unstructured": "GPUTreeShap: massively parallel exact calculation of SHAP scores for tree ensembles [PeerJ]. Accessed August 18, 2022. https://peerj.com/articles/cs-880/"
},
{
"key": "2022122802000890000_2022.12.22.22283791v1.20",
"unstructured": "XGBoost | Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. Accessed August 18, 2022. https://dl.acm.org/doi/10.1145/2939672.2939785"
},
{
"DOI": "10.1007/s00431-022-04600-x",
"doi-asserted-by": "publisher",
"key": "2022122802000890000_2022.12.22.22283791v1.21"
}
],
"reference-count": 21,
"references-count": 21,
"relation": {},
"resource": {
"primary": {
"URL": "http://medrxiv.org/lookup/doi/10.1101/2022.12.22.22283791"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subtitle": [],
"subtype": "preprint",
"title": "A machine learning-based phenotype for long COVID in children: an EHR-based study from the RECOVER program",
"type": "posted-content"
}