Distal Protein-Protein Interactions Contribute to SARS-CoV-2 Main Protease Substrate Binding and Nirmatrelvir Resistance
et al., bioRxiv, doi:10.1101/2024.04.01.587566, Apr 2024
In vitro and crystallographic study reveals that the L50F mutation in SARS-CoV-2 main protease (Mpro) can restore the reduced enzymatic activity caused by nirmatrelvir resistance mutations E166A/L167F through enhanced protein-protein interactions outside the active site. The L50F/E166A/L167F triple mutant exhibited near wild-type activity with Mpro protein substrates, suggesting resistance to paxlovid. The study highlights the importance of considering distal mutations when assessing drug resistance and the need for comprehensive variant surveillance.
Lewandowski et al., 2 Apr 2024, multiple countries, preprint, 10 authors.
Contact: ychen1@usf.edu, junwang@pharmacy.rutgers.edu.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Distal Protein-Protein Interactions Contribute to SARS-CoV-2 Main Protease Substrate Binding and Nirmatrelvir Resistance
doi:10.1101/2024.04.01.587566
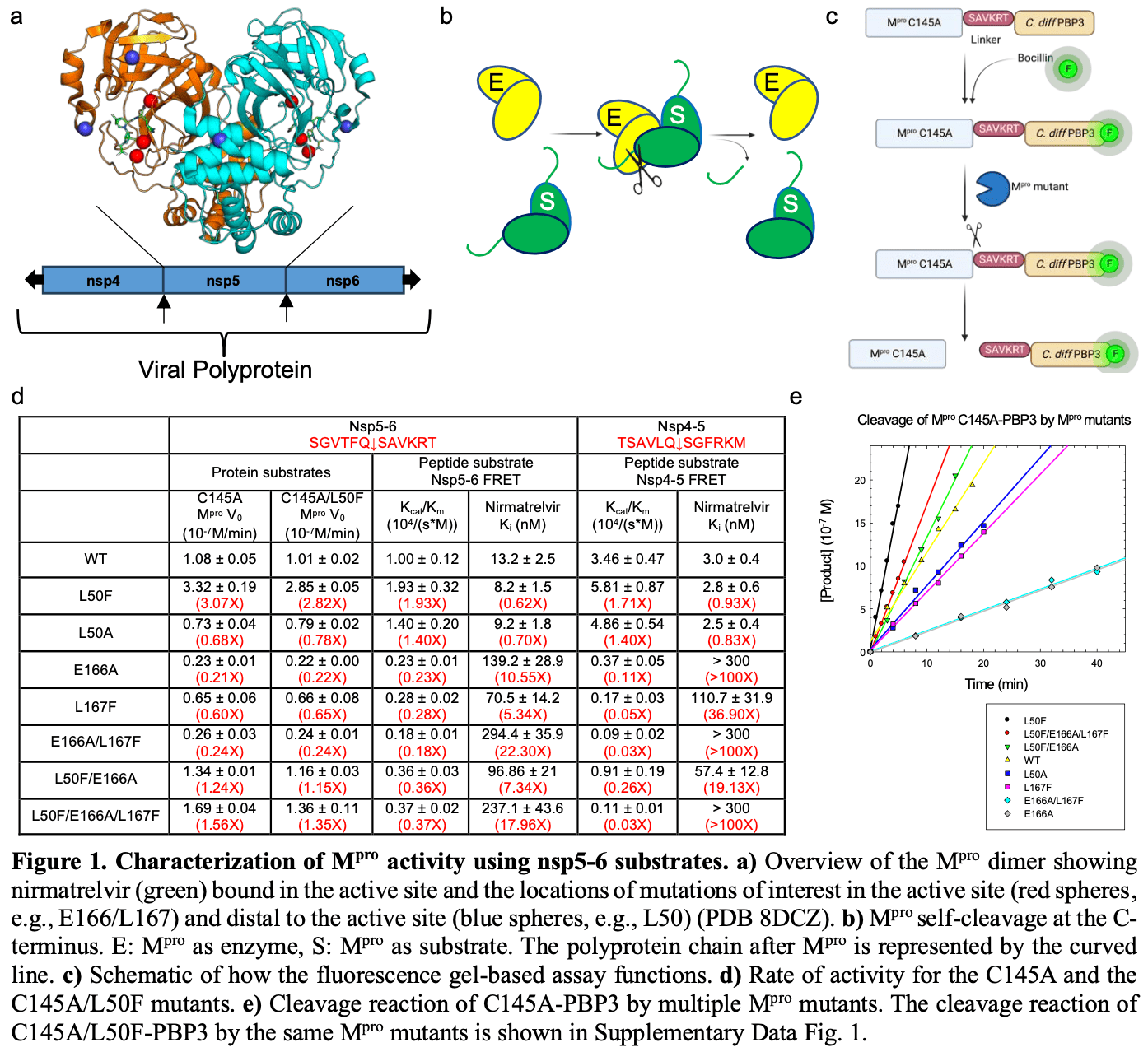
SARS-CoV-2 main protease, M pro , is responsible for the processing of the viral polyproteins into individual proteins, including the protease itself. M pro is a key target of anti-COVID-19 therapeutics such as nirmatrelvir (the active component of Paxlovid). Resistance mutants identified clinically and in viral passage assays contain a combination of active site mutations (e.g. E166V, E166A, L167F), which reduce inhibitor binding and enzymatic activity, and non-active site mutations (e.g. P252L, T21I, L50F), which restore the fitness of viral replication. Although the mechanism of resistance for the active site mutations is apparent, the role of the non-active site mutations in fitness rescue remains elusive. In this study, we use the model system of a M pro triple mutant (L50F/E166A/L167F) that confers not only nirmatrelvir drug resistance but also a similar fitness of replication compared to the wild-type both in vitro and in vivo. By comparing peptide and full-length M pro protein as substrates, we demonstrate that the binding of M pro substrate involves more than residues in the active site. In particular, L50F and other non-active site mutations can enhance the M pro dimer-dimer interactions and help place the nsp5-6 substrate at the enzyme catalytic center. The structural and enzymatic activity data of M pro L50F, L50F/E166A/L167F, and others underscore the importance of considering the whole substrate protein in studying M pro and substrate interactions, and offers important insights into M pro function, resistance development, and inhibitor design.
Author Contributions: E.M.L, J.W, and Y.C wrote the manuscript; X.Z, A.F, and L.M.C.J carried out the gel-assay; H.T. performed the FRET enzymatic assay; E.M.L and A.J.B crystallized proteins; E.M.L and N.K was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
Competing Interests: The authors declare no competing financial interests.
References
Abdelnabi, Nirmatrelvir-resistant SARS-CoV-2 is efficiently transmitted in female Syrian hamsters and retains partial susceptibility to treatment, Nat Commun
Aniana, Insights into the mechanism of SARS-CoV-2 main protease autocatalytic maturation from model precursors, Commun Biol
Cady, Wang, Wu, Degrado, Hong, Specific binding of adamantane drugs and direction of their polar amines in the pore of the influenza M2 transmembrane domain in lipid bilayers and dodecylphosphocholine micelles determined by NMR spectroscopy, J Am Chem Soc
Chen, SARS-CoV-2 M(pro) Protease Variants of Concern Display Altered Viral Substrate and Cell Host Target Galectin-8 Processing but Retain Sensitivity toward Antivirals, ACS Cent Sci
Duan, Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir, Nature
Emsley, Cowtan, Coot: model-building tools for molecular graphics, Acta Cryst D Biol Crystallog
Hu, Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir, ACS Cent Sci
Humphrey, Dalke, Schulten, VMD: visual molecular dynamics, J Mol Graph
Iketani, Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir, Nature
Jin, Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors, Nature
Jochmans, The Substitutions L50F, E166A, and L167F in SARS-CoV-2 3CLpro Are Selected by a Protease Inhibitor In Vitro and Confer Resistance To Nirmatrelvir, mBio
Joosten, Long, Murshudov, Perrakis, The PDB_REDO server for macromolecular structure model optimization, IUCrJ
Joyce, Hu, Wang, The history, mechanism, and perspectives of nirmatrelvir (PF-07321332): an orally bioavailable main protease inhibitor used in combination with ritonavir to reduce COVID-19-related hospitalizations, Med Chem Res
Kiso, In vitro and in vivo characterization of SARS-CoV-2 strains resistant to nirmatrelvir, Nat Commun
Najjar-Debbiny, Effectiveness of Paxlovid in Reducing Severe Coronavirus Disease 2019 and Mortality in High-Risk Patients, Clin Infect Dis
Owen, An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19, Science
Sanner, Olson, Spehner, Methods: Fluorescence gel assay fusion protein construct and purification The M pro C145A (C145A/L50F) and C. difficile PBP3 42-554 fusion protein was inserted into the pETGSTSumo vector. The cleavage site, SAVKRT, was inserted between M pro and PBP3 42-554. The expression constructs were transformed into Rosetta (DE3) pLysS cells. A single colony was picked and grew in LB (Luria-Bertani) media supplemented with 50 µg/mL kanamycin and 35 µg/mL chloramphenicol at 37 °C overnight. The overnight culture was then added into 1 L media at 1:100 and incubated at 37 °C until the OD600 reached 0.4. Protein expression was induced using, Biopolymers
Shaqra, Defining the substrate envelope of SARS-CoV-2 main protease to predict and avoid drug resistance, Nat Commun
Zhou, Nirmatrelvir-resistant SARS-CoV-2 variants with high fitness in an infectious cell culture system, Sci Adv
Zuckerman, Bucris, Keidar-Friedman, Amsalem, Brosh-Nissimov, Nirmatrelvir Resistance-de Novo E166V/L50V Mutations in an Immunocompromised Patient Treated With Prolonged Nirmatrelvir/Ritonavir Monotherapy Leading to Clinical and Virological Treatment Failure-a Case Report, Clin Infect Dis
DOI record:
{
"DOI": "10.1101/2024.04.01.587566",
"URL": "http://dx.doi.org/10.1101/2024.04.01.587566",
"abstract": "<jats:title>Abstract</jats:title><jats:p>SARS-CoV-2 main protease, M<jats:sup>pro</jats:sup>, is responsible for the processing of the viral polyproteins into individual proteins, including the protease itself. M<jats:sup>pro</jats:sup>is a key target of anti-COVID-19 therapeutics such as nirmatrelvir (the active component of Paxlovid). Resistance mutants identified clinically and in viral passage assays contain a combination of active site mutations (e.g. E166V, E166A, L167F), which reduce inhibitor binding and enzymatic activity, and non-active site mutations (e.g. P252L, T21I, L50F), which restore the fitness of viral replication. Although the mechanism of resistance for the active site mutations is apparent, the role of the non-active site mutations in fitness rescue remains elusive. In this study, we use the model system of a M<jats:sup>pro</jats:sup>triple mutant (L50F/E166A/L167F) that confers not only nirmatrelvir drug resistance but also a similar fitness of replication compared to the wild-type both in vitro and in vivo. By comparing peptide and full-length M<jats:sup>pro</jats:sup>protein as substrates, we demonstrate that the binding of M<jats:sup>pro</jats:sup>substrate involves more than residues in the active site. In particular, L50F and other non-active site mutations can enhance the M<jats:sup>pro</jats:sup>dimer-dimer interactions and help place the nsp5-6 substrate at the enzyme catalytic center. The structural and enzymatic activity data of M<jats:sup>pro</jats:sup>L50F, L50F/E166A/L167F, and others underscore the importance of considering the whole substrate protein in studying M<jats:sup>pro</jats:sup>and substrate interactions, and offers important insights into M<jats:sup>pro</jats:sup>function, resistance development, and inhibitor design.</jats:p>",
"accepted": {
"date-parts": [
[
2024,
4,
2
]
]
},
"author": [
{
"affiliation": [],
"family": "Lewandowski",
"given": "Eric M.",
"sequence": "first"
},
{
"affiliation": [],
"family": "Zhang",
"given": "Xiujun",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Tan",
"given": "Haozhou",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Jaskolka-Brown",
"given": "Aiden",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Kohaal",
"given": "Navita",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Frazier",
"given": "Aliaksandra",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-1411-9080",
"affiliation": [],
"authenticated-orcid": false,
"family": "Madsen",
"given": "Jesper J.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Jacobs",
"given": "Lian M.C.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-4845-4621",
"affiliation": [],
"authenticated-orcid": false,
"family": "Wang",
"given": "Jun",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Chen",
"given": "Yu",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
4,
3
]
],
"date-time": "2024-04-03T02:40:12Z",
"timestamp": 1712112012000
},
"deposited": {
"date-parts": [
[
2024,
4,
5
]
],
"date-time": "2024-04-05T14:25:39Z",
"timestamp": 1712327139000
},
"group-title": "Molecular Biology",
"indexed": {
"date-parts": [
[
2024,
4,
6
]
],
"date-time": "2024-04-06T00:48:16Z",
"timestamp": 1712364496369
},
"institution": [
{
"name": "bioRxiv"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2024,
4,
2
]
]
},
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2024.04.01.587566",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2024,
4,
2
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2024,
4,
2
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference": [
{
"DOI": "10.1001/jama.2023.27841",
"article-title": "As COVID-19 Cases Surge, Here’s What to Know About JN.1, the Latest SARS-CoV-2 \"Variant of Interest\"",
"doi-asserted-by": "crossref",
"first-page": "382",
"journal-title": "Jama",
"key": "2024040507251817000_2024.04.01.587566v1.1",
"volume": "331",
"year": "2024"
},
{
"article-title": "The SARS-CoV-2 main protease (M(pro)): Structure, function, and emerging therapies for COVID-19",
"first-page": "e151",
"journal-title": "MedComm (2020)",
"key": "2024040507251817000_2024.04.01.587566v1.2",
"volume": "3",
"year": "2022"
},
{
"DOI": "10.1038/s41586-020-2223-y",
"article-title": "Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors",
"doi-asserted-by": "crossref",
"first-page": "289",
"journal-title": "Nature",
"key": "2024040507251817000_2024.04.01.587566v1.3",
"volume": "582",
"year": "2020"
},
{
"DOI": "10.1126/science.abl4784",
"article-title": "An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19",
"doi-asserted-by": "crossref",
"first-page": "1586",
"journal-title": "Science (New York, N.Y.)",
"key": "2024040507251817000_2024.04.01.587566v1.4",
"volume": "374",
"year": "2021"
},
{
"DOI": "10.1007/s00044-022-02951-6",
"doi-asserted-by": "publisher",
"key": "2024040507251817000_2024.04.01.587566v1.5"
},
{
"DOI": "10.1093/cid/ciac443",
"article-title": "Effectiveness of Paxlovid in Reducing Severe Coronavirus Disease 2019 and Mortality in High-Risk Patients",
"doi-asserted-by": "crossref",
"first-page": "e342",
"journal-title": "Clin Infect Dis",
"key": "2024040507251817000_2024.04.01.587566v1.6",
"volume": "76",
"year": "2023"
},
{
"DOI": "10.1021/acscentsci.3c00538",
"article-title": "Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir",
"doi-asserted-by": "crossref",
"first-page": "1658",
"journal-title": "ACS Cent Sci",
"key": "2024040507251817000_2024.04.01.587566v1.7",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.1038/s41586-022-05514-2",
"article-title": "Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir",
"doi-asserted-by": "crossref",
"first-page": "558",
"journal-title": "Nature",
"key": "2024040507251817000_2024.04.01.587566v1.8",
"volume": "613",
"year": "2023"
},
{
"DOI": "10.1128/mbio.02815-22",
"article-title": "The Substitutions L50F, E166A, and L167F in SARS-CoV-2 3CLpro Are Selected by a Protease Inhibitor In Vitro and Confer Resistance To Nirmatrelvir",
"doi-asserted-by": "crossref",
"first-page": "e0281522",
"journal-title": "mBio",
"key": "2024040507251817000_2024.04.01.587566v1.9",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1038/s41467-023-37773-6",
"article-title": "Nirmatrelvir-resistant SARS-CoV-2 is efficiently transmitted in female Syrian hamsters and retains partial susceptibility to treatment",
"doi-asserted-by": "crossref",
"first-page": "2124",
"journal-title": "Nat Commun",
"key": "2024040507251817000_2024.04.01.587566v1.10",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1038/s41467-023-39704-x",
"doi-asserted-by": "publisher",
"key": "2024040507251817000_2024.04.01.587566v1.11"
},
{
"DOI": "10.1021/acscentsci.3c00054",
"article-title": "SARS-CoV-2 M(pro) Protease Variants of Concern Display Altered Viral Substrate and Cell Host Target Galectin-8 Processing but Retain Sensitivity toward Antivirals",
"doi-asserted-by": "crossref",
"first-page": "696",
"journal-title": "ACS Cent Sci",
"key": "2024040507251817000_2024.04.01.587566v1.12",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.1126/sciadv.add7197",
"article-title": "Nirmatrelvir-resistant SARS-CoV-2 variants with high fitness in an infectious cell culture system",
"doi-asserted-by": "crossref",
"first-page": "eadd7197",
"journal-title": "Sci Adv",
"key": "2024040507251817000_2024.04.01.587566v1.13",
"volume": "8",
"year": "2022"
},
{
"article-title": "Nirmatrelvir Resistance—de Novo E166V/L50V Mutations in an Immunocompromised Patient Treated With Prolonged Nirmatrelvir/Ritonavir Monotherapy Leading to Clinical and Virological Treatment Failure—a Case Report",
"first-page": "352",
"journal-title": "Clin Infect Dis",
"key": "2024040507251817000_2024.04.01.587566v1.14",
"volume": "78",
"year": "2023"
},
{
"DOI": "10.1038/s41586-023-06609-0",
"article-title": "Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir",
"doi-asserted-by": "crossref",
"first-page": "376",
"journal-title": "Nature",
"key": "2024040507251817000_2024.04.01.587566v1.15",
"volume": "622",
"year": "2023"
},
{
"DOI": "10.1038/s42003-023-05469-8",
"article-title": "Insights into the mechanism of SARS-CoV-2 main protease autocatalytic maturation from model precursors",
"doi-asserted-by": "crossref",
"first-page": "1159",
"journal-title": "Commun Biol",
"key": "2024040507251817000_2024.04.01.587566v1.16",
"volume": "6",
"year": "2023"
},
{
"DOI": "10.1038/s41467-022-31210-w",
"doi-asserted-by": "publisher",
"key": "2024040507251817000_2024.04.01.587566v1.17"
},
{
"DOI": "10.1073/pnas.2117142119",
"doi-asserted-by": "publisher",
"key": "2024040507251817000_2024.04.01.587566v1.18"
},
{
"DOI": "10.1038/s41467-020-19662-4",
"article-title": "Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site",
"doi-asserted-by": "crossref",
"first-page": "5877",
"journal-title": "Nat Commun",
"key": "2024040507251817000_2024.04.01.587566v1.19",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1021/ja102581n",
"doi-asserted-by": "publisher",
"key": "2024040507251817000_2024.04.01.587566v1.20"
},
{
"DOI": "10.1107/S0907444994003112",
"article-title": "The CCP4 suite: programs for protein crystallography",
"author": "Collaborative Computational Project, N",
"doi-asserted-by": "crossref",
"first-page": "760",
"journal-title": "Acta Cryst D Biol Crystallog",
"key": "2024040507251817000_2024.04.01.587566v1.21",
"volume": "50",
"year": "1994"
},
{
"DOI": "10.1107/S0907444904019158",
"article-title": "Coot: model-building tools for molecular graphics",
"doi-asserted-by": "crossref",
"first-page": "2126",
"journal-title": "Acta Cryst D Biol Crystallog",
"key": "2024040507251817000_2024.04.01.587566v1.22",
"volume": "60",
"year": "2004"
},
{
"DOI": "10.1107/S2052252514009324",
"doi-asserted-by": "publisher",
"key": "2024040507251817000_2024.04.01.587566v1.23"
},
{
"DOI": "10.1016/0263-7855(96)00018-5",
"doi-asserted-by": "publisher",
"key": "2024040507251817000_2024.04.01.587566v1.24"
},
{
"DOI": "10.1002/(SICI)1097-0282(199603)38:3<305::AID-BIP4>3.0.CO;2-Y",
"doi-asserted-by": "publisher",
"key": "2024040507251817000_2024.04.01.587566v1.25"
}
],
"reference-count": 25,
"references-count": 25,
"relation": {},
"resource": {
"primary": {
"URL": "http://biorxiv.org/lookup/doi/10.1101/2024.04.01.587566"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subtitle": [],
"subtype": "preprint",
"title": "Distal Protein-Protein Interactions Contribute to SARS-CoV-2 Main Protease Substrate Binding and Nirmatrelvir Resistance",
"type": "posted-content"
}