Computational prediction of interactions between Paxlovid and prescription drugs
et al., Proceedings of the National Academy of Sciences, doi:10.1073/pnas.2221857120, Mar 2023
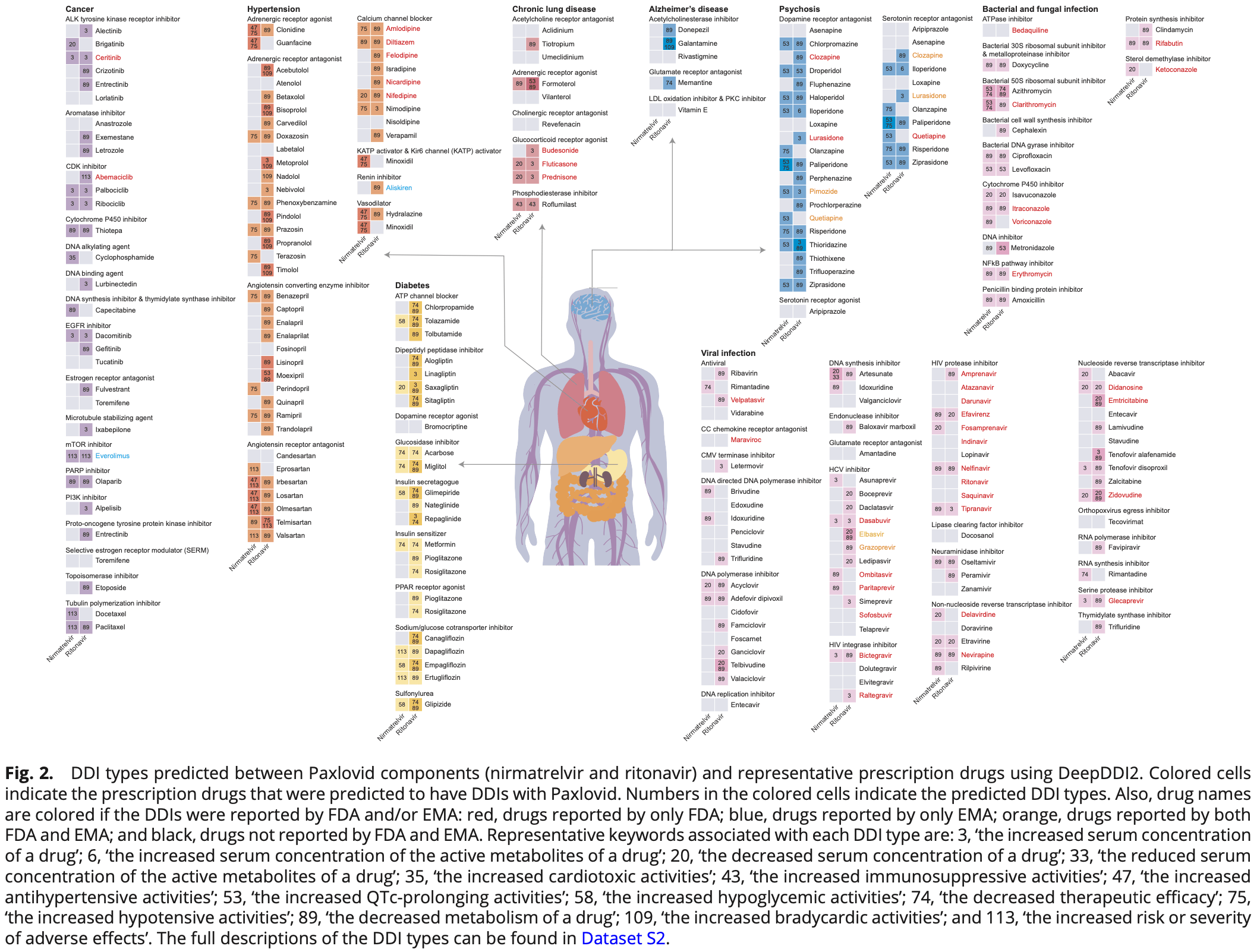
In silico analysis of drug-drug interactions for paxlovid. From 2,248 prescription drugs, 1,628 were predicted to have 2,445 interactions with nirmatrelvir and/or ritonavir (673 for nirmatrelvir and 1,403 ritonavir). For 873 drugs, authors provide a list of possible alternatives that share mechanisms of action, but are predicted to have fewer or no interactions.
Kim et al., 13 Mar 2023, peer-reviewed, 4 authors.
Contact: leesy@kaist.ac.kr.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Abstract: BRIEF REPORT
| BIOPHYSICS AND COMPUTATIONAL BIOLOGY
OPEN ACCESS
Computational prediction of interactions between Paxlovid
and prescription drugs
Yeji Kima,b,1
, Jae Yong Ryuc,1
, Hyun Uk Kima,b,d,e,1
, and Sang Yup Leea,b,e,2
Edited by Jens Nielsen, BioInnovation Institute, Copenhagen, Denmark; received December 29, 2022; accepted February 14, 2023
Pfizer’s Paxlovid has recently been approved for the emergency use authorization
(EUA) from the US Food and Drug Administration (FDA) for the treatment of
mild-to-moderate COVID-19. Drug interactions can be a serious medical problem
for COVID-19 patients with underlying medical conditions, such as hypertension
and diabetes, who have likely been taking other drugs. Here, we use deep learning to
predict potential drug–drug interactions between Paxlovid components (nirmatrelvir
and ritonavir) and 2,248 prescription drugs for treating various diseases.
COVID-19 | drug interactions | DeepDDI2 | Paxlovid
In December 2021, Pfizer’s Paxlovid (nirmatrelvir and ritonavir copackaged for oral use)
received Emergency Use Authorization (EUA) from the US Food and Drug Administration
(FDA) for the treatment of mild-to-moderate COVID-19 patients. Nirmatrelvir inhibits
the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) 3C-like protease to
prevent virus replication, and ritonavir slows down the degradation of nirmatrelvir by
acting as a CYP3A inhibitor. Subsequent clinical studies showed that Paxlovid is effective
in reducing the hospitalization risk of COVID-19 patients aged 50 or older (1). A similar
result was observed in patients aged 65 or older, who were treated with nirmatrelvir alone
(2). Importantly, when the EUA was issued, FDA also provided information on 108 drugs
that might exhibit potential drug interactions with Paxlovid (3). Likewise, in January
2022, European Medicines Agency (EMA) also reported 128 drugs that can potentially
interact with Paxlovid (‘Paxlovid: EPAR—Product information’ available at: https://www.
ema.europa.eu/en/medicines/human/EPAR/paxlovid#product-information-section;
Accessed December 22, 2022); FDA and EMA reported 69 drugs in common. Both FDA
and EMA reported potentially interacting drugs mainly because these drugs can serve as
substrates or inhibitors of cytochromes P450 (CYPs), leading to unwanted drug–drug interactions (DDIs). Such DDIs can be a serious problem for the COVID-19 patients having
underlying medical conditions such as hypertension and diabetes because these patients are
already taking medicine to treat their conditions (4–7). A problem here is that there are
likely more drugs that might interact with Paxlovid, and possible DDIs involving Paxlovid
cannot be experimentally examined in a short period of time.
Here, we report the list of a large number of prescription drugs that are predicted to have
DDIs with Paxlovid by employing DeepDDI. DeepDDI is a computational model developed
using deep learning that predicts the pharmacological effects and adverse drug events (ADEs)
of DDIs (8). DeepDDI receives structural information as simplified molecular-input line-entry
system of two drugs in a pair as an input, and predicts DDI types as an output in the form of
human-readable sentences. The DeepDDI output sentences describe changes in the pharmacological effects and/or the risk of ADEs as a result of the DDI. DeepDDI originally..
DOI record:
{
"DOI": "10.1073/pnas.2221857120",
"ISSN": [
"0027-8424",
"1091-6490"
],
"URL": "http://dx.doi.org/10.1073/pnas.2221857120",
"abstract": "<jats:p>Pfizer’s Paxlovid has recently been approved for the emergency use authorization (EUA) from the US Food and Drug Administration (FDA) for the treatment of mild-to-moderate COVID-19. Drug interactions can be a serious medical problem for COVID-19 patients with underlying medical conditions, such as hypertension and diabetes, who have likely been taking other drugs. Here, we use deep learning to predict potential drug–drug interactions between Paxlovid components (nirmatrelvir and ritonavir) and 2,248 prescription drugs for treating various diseases.</jats:p>",
"alternative-id": [
"10.1073/pnas.2221857120"
],
"assertion": [
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2022-12-29"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 1,
"value": "2023-02-14"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 2,
"value": "2023-03-13"
}
],
"author": [
{
"ORCID": "http://orcid.org/0000-0001-9801-917X",
"affiliation": [
{
"name": "Metabolic and Biomolecular Engineering National Research Laboratory, Department of Chemical and Biomolecular Engineering (BK21 four), Korea Advanced Institute of Science and Technology, Daejeon 34141, Republic of Korea"
},
{
"name": "Systems Metabolic Engineering and Systems Healthcare Cross-Generation Collaborative Laboratory, Korea Advanced Institute of Science and Technology, Daejeon 34141, Republic of Korea"
}
],
"authenticated-orcid": false,
"family": "Kim",
"given": "Yeji",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0003-0603-1599",
"affiliation": [
{
"name": "Department of Biotechnology, Duksung Women’s University, Seoul 01369, Republic of Korea"
}
],
"authenticated-orcid": false,
"family": "Ryu",
"given": "Jae Yong",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-7224-642X",
"affiliation": [
{
"name": "Metabolic and Biomolecular Engineering National Research Laboratory, Department of Chemical and Biomolecular Engineering (BK21 four), Korea Advanced Institute of Science and Technology, Daejeon 34141, Republic of Korea"
},
{
"name": "Systems Metabolic Engineering and Systems Healthcare Cross-Generation Collaborative Laboratory, Korea Advanced Institute of Science and Technology, Daejeon 34141, Republic of Korea"
},
{
"name": "Systems Biology and Medicine Laboratory, Department of Chemical and Biomolecular Engineering, Korea Advanced Institute of Science and Technology, Daejeon 34141, Republic of Korea"
},
{
"name": "KAIST Institute for the BioCentury, BioProcess Engineering Research Center, and BioInformatics Research Center, Korea Advanced Institute of Science and Technology, Daejeon 34141, Republic of Korea"
}
],
"authenticated-orcid": false,
"family": "Kim",
"given": "Hyun Uk",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-0599-3091",
"affiliation": [
{
"name": "Metabolic and Biomolecular Engineering National Research Laboratory, Department of Chemical and Biomolecular Engineering (BK21 four), Korea Advanced Institute of Science and Technology, Daejeon 34141, Republic of Korea"
},
{
"name": "Systems Metabolic Engineering and Systems Healthcare Cross-Generation Collaborative Laboratory, Korea Advanced Institute of Science and Technology, Daejeon 34141, Republic of Korea"
},
{
"name": "KAIST Institute for the BioCentury, BioProcess Engineering Research Center, and BioInformatics Research Center, Korea Advanced Institute of Science and Technology, Daejeon 34141, Republic of Korea"
}
],
"authenticated-orcid": false,
"family": "Lee",
"given": "Sang Yup",
"sequence": "additional"
}
],
"container-title": "Proceedings of the National Academy of Sciences",
"container-title-short": "Proc. Natl. Acad. Sci. U.S.A.",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"www.pnas.org"
]
},
"created": {
"date-parts": [
[
2023,
3,
13
]
],
"date-time": "2023-03-13T19:11:16Z",
"timestamp": 1678734676000
},
"deposited": {
"date-parts": [
[
2023,
3,
13
]
],
"date-time": "2023-03-13T19:12:10Z",
"timestamp": 1678734730000
},
"funder": [
{
"DOI": "10.13039/501100014188",
"award": [
"MCM-2020-N11200225"
],
"doi-asserted-by": "publisher",
"name": "Ministry of Science and ICT, South Korea"
}
],
"indexed": {
"date-parts": [
[
2023,
3,
14
]
],
"date-time": "2023-03-14T04:54:17Z",
"timestamp": 1678769657182
},
"is-referenced-by-count": 0,
"issue": "12",
"issued": {
"date-parts": [
[
2023,
3,
13
]
]
},
"journal-issue": {
"issue": "12",
"published-print": {
"date-parts": [
[
2023,
3,
21
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
3,
13
]
],
"date-time": "2023-03-13T00:00:00Z",
"timestamp": 1678665600000
}
}
],
"link": [
{
"URL": "https://pnas.org/doi/pdf/10.1073/pnas.2221857120",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "341",
"original-title": [],
"prefix": "10.1073",
"published": {
"date-parts": [
[
2023,
3,
13
]
]
},
"published-online": {
"date-parts": [
[
2023,
3,
13
]
]
},
"published-print": {
"date-parts": [
[
2023,
3,
21
]
]
},
"publisher": "Proceedings of the National Academy of Sciences",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "https://pnas.org/doi/10.1073/pnas.2221857120"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Multidisciplinary"
],
"subtitle": [],
"title": "Computational prediction of interactions between Paxlovid and prescription drugs",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1073/pnas.cm10313",
"volume": "120"
}