Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease
et al., Life Sciences, doi:10.1016/j.lfs.2020.117627, Jun 2020
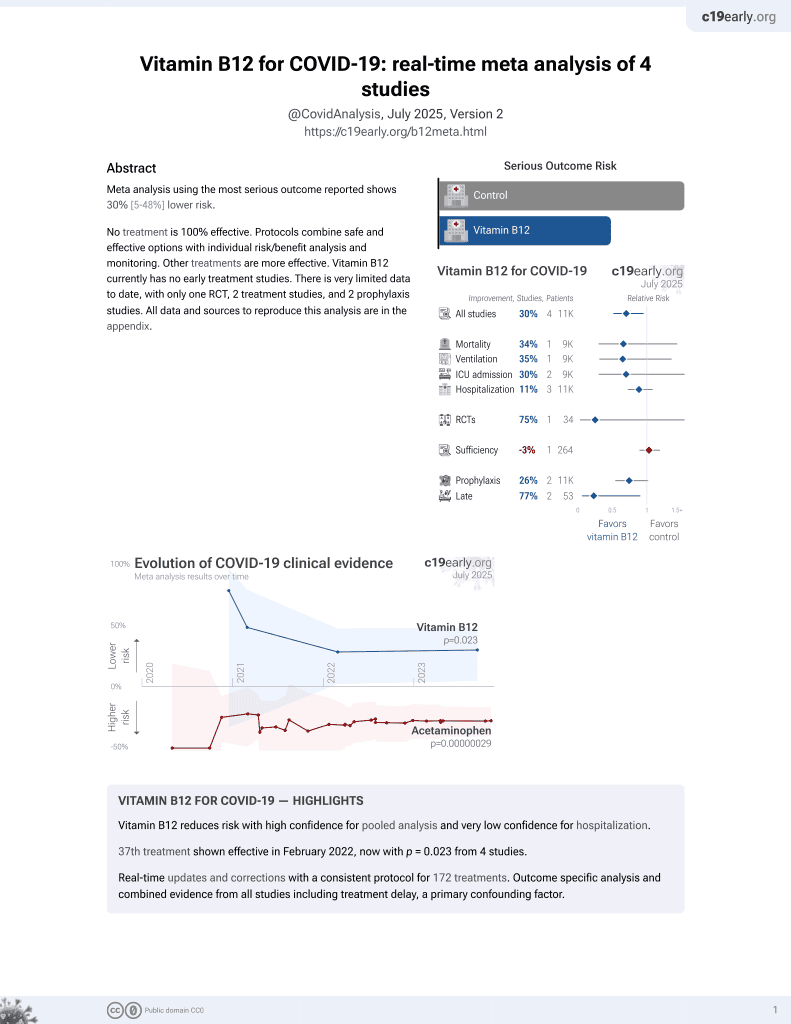
38th treatment shown to reduce risk in
February 2022, now with p = 0.023 from 4 studies.
Lower risk for recovery.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
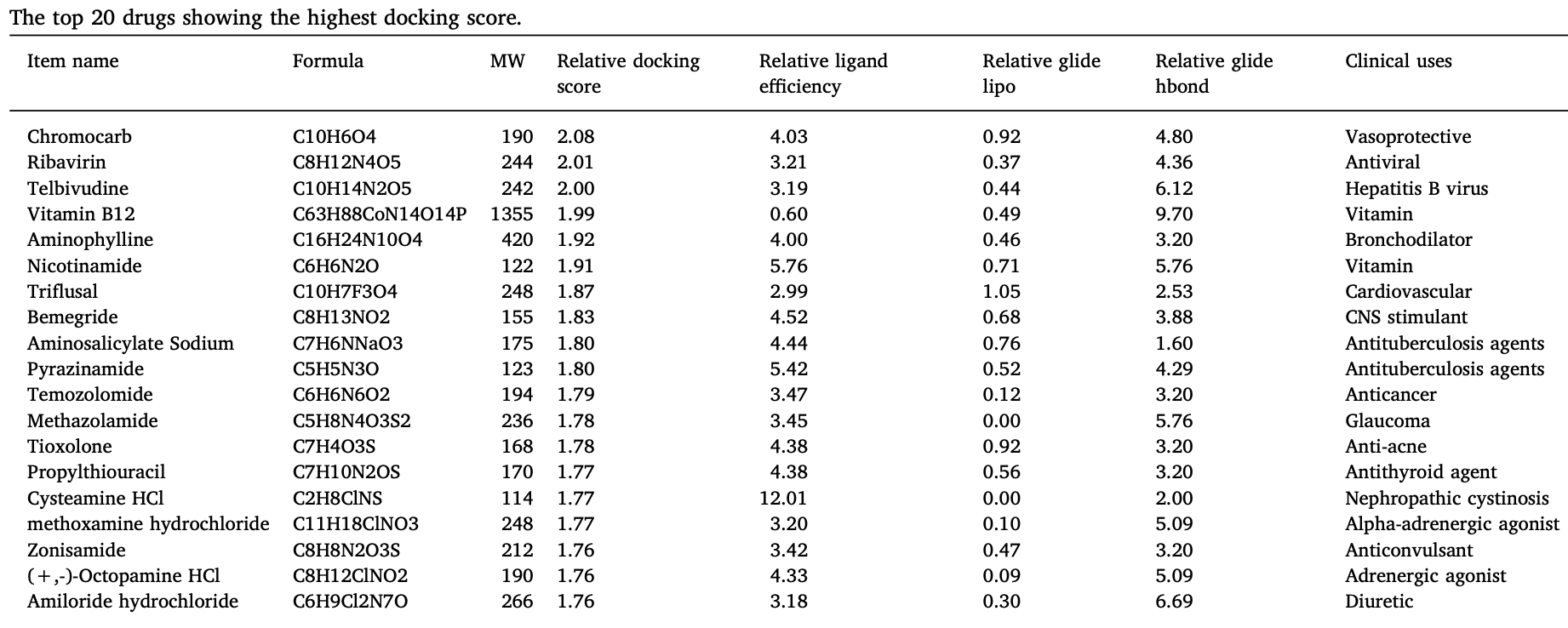
In silico study of 1,500 FDA-approved drugs finding that vitamin B12 had strong binding affinity with SARS-CoV-2 Mpro, and may be beneficial for COVID-19 treatment.
Kandeel et al., 30 Jun 2020, China, peer-reviewed, 2 authors.
Contact: mkandeel@kfu.edu.sa.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease
Life Sciences, doi:10.1016/j.lfs.2020.117627
Aims: In December 2019, the Coronavirus disease-2019 (COVID-19) virus has emerged in Wuhan, China. In this research, the first resolved COVID-19 crystal structure (main protease) was targeted in a virtual screening study by of FDA approved drugs dataset. In addition, a knowledge gap in relations of COVID-19 with the previously known fatal Coronaviruses (CoVs) epidemics, SARS and MERS CoVs, was covered by investigation of sequence statistics and phylogenetics. Materials and methods: Molecular modeling, virtual screening, docking, sequence comparison statistics and phylogenetics of the COVID-19 main protease were investigated. Key findings: COVID-19 Mpro formed a phylogenetic group with SARS CoV that was distant from MERS CoV. The identity% was 96.061 and 51.61 for COVID-19/SARS and COVID-19/MERS CoV sequence comparisons, respectively. The top 20 drugs in the virtual screening studies comprised a broad-spectrum antiviral (ribavirin), anti-hepatitis B virus (telbivudine), two vitamins (vitamin B12 and nicotinamide) and other miscellaneous systemically acting drugs. Of special interest, ribavirin had been used in treating cases of SARS CoV. Significance: The present study provided a comprehensive targeting of the first resolved COVID+19 structure of Mpro and found a suitable save drugs for repurposing against the viral Mpro. Ribavirin, telbivudine, vitamin B12 and nicotinamide can be combined and used for COVID treatment. This initiative relocates already marketed and approved safe drugs for potential use in COVID-treatment.
Declaration of competing interest The authors declare no conflict of interest.
References
Altaher, Kandeel, Molecular analysis of some camel cytochrome P450 enzymes reveals lower evolution and drug-binding properties, J. Biomol. Struct. Dyn
Altaher, Nakanishi, Kandeel, Annotation of camel genome for estimation of drug binding power, evolution and adaption of cytochrome P450 1a2, Int. J. Pharmacol
Crotty, Maag, Arnold, Zhong, Lau et al., The broadspectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen, Nat. Med
Desenclos, Van Der Werf, Bonmarin, Levy-Bruhl, Yazdanpanah et al., Introduction of SARS in France, Emerg. Infect. Dis
Elhefnawi, Elgamacy, Fares, Multiple virtual screening approaches for finding new Hepatitis c virus RNA-dependent RNA polymerase inhibitors: structurebased screens and molecular dynamics for the pursue of new poly pharmacological inhibitors, BMC Bioinf, BioMed Central
Friesner, Jay, Murphy, Halgren, Klicic et al., Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy, J. Med. Chem
Guarner, Three emerging coronaviruses in two decades: the story of SARS, MERS, and now COVID-19, Am. J. Clin. Pathol, doi:10.1093/ajcp/aqaa029
Halgren, Murphy, Friesner, Beard, Frye et al., Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening, J. Med. Chem
Hilgenfeld, From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design, FEBS J
Kandeel, Altaher, Alnazawi, Molecular dynamics and inhibition of MERS CoV papain-like protease by small molecule imidazole and aminopurine derivatives, Lett. Drug Des. Discov
Kandeel, Ando, Kitamura, Abdel-Aziz, Kitade, Mutational, inhibitory and microcalorimetric analyses of Plasmodium falciparum TMP kinase. Implications for drug discovery, Parasitology
Koren, King, Knowles, Phillips, Ribavirin in the treatment of SARS: a new trick for an old drug?, CMAJ
Li, Hu, Wu, Yao, Li, Molecular characteristics, functions, and related pathogenicity of MERS-CoV proteins, Engineering, doi:10.1016/j.eng.2018.11.035
Li, Wong, Li, Kuhn, Huang et al., Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions, J. Virol
Liu, Zhou, SARS-CoV protease inhibitors design using virtual screening method from natural products libraries, J. Comput. Chem
Mckeage, Keam, None, Telbivudine
Murgueitio, Bermudez, Mortier, Wolber, In silico virtual screening approaches for anti-viral drug discovery, Drug Discov. Today Technol
Peiris, Lai, Poon, Guan, Yam et al., Coronavirus as a possible cause of severe acute respiratory syndrome, Lancet
Pillaiyar, Meenakshisundaram, Manickam, Recent discovery and development of inhibitors targeting coronaviruses, Drug Discov. Today
Raj, Varadwaj, Flavonoids as multi-target inhibitors for proteins associated with Ebola virus: in silico discovery using virtual screening and molecular docking studies, Interdiscip. Sci. Comput. Life Sci
Sheikh, Al-Taher, Al-Nazawi, Al-Mubarak, Kandeel, Analysis of preferred codon usage in the coronavirus N genes and their implications for genome evolution and vaccine design, J. Virol. Methods
Sirois, Wei, Du, Chou, Virtual screening for SARS-CoV protease based on KZ7088 pharmacophore points, J. Chem. Inf. Comput. Sci
Wan, Shang, Graham, Baric, Li, Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS, J. Virol, doi:10.1128/JVI.00127-20
Wen, Kuo, Jan, Liang, Wang et al., Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus, J. Med. Chem
Xu, Chen, Wang, Feng, Zhou et al., Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission, Sci. China Life Sci
Zaki, Van Boheemen, Bestebroer, Osterhaus, Fouchier, Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia, N. Engl. J. Med
Zhou, Khaliq, Suk, Patkar, Li et al., Antiviral compounds discovered by virtual screening of small-molecule libraries against dengue virus E protein, ACS Chem. Biol
Zumla, Chan, Azhar, Hui, Yuen, Coronaviruses-drug discovery and therapeutic options, Nat. Rev. Drug Discov
DOI record:
{
"DOI": "10.1016/j.lfs.2020.117627",
"ISSN": [
"0024-3205"
],
"URL": "http://dx.doi.org/10.1016/j.lfs.2020.117627",
"alternative-id": [
"S0024320520303751"
],
"article-number": "117627",
"assertion": [
{
"label": "This article is maintained by",
"name": "publisher",
"value": "Elsevier"
},
{
"label": "Article Title",
"name": "articletitle",
"value": "Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease"
},
{
"label": "Journal Title",
"name": "journaltitle",
"value": "Life Sciences"
},
{
"label": "CrossRef DOI link to publisher maintained version",
"name": "articlelink",
"value": "https://doi.org/10.1016/j.lfs.2020.117627"
},
{
"label": "Content Type",
"name": "content_type",
"value": "article"
},
{
"label": "Copyright",
"name": "copyright",
"value": "© 2020 Elsevier Inc. All rights reserved."
}
],
"author": [
{
"affiliation": [],
"family": "Kandeel",
"given": "Mahmoud",
"sequence": "first"
},
{
"affiliation": [],
"family": "Al-Nazawi",
"given": "Mohammed",
"sequence": "additional"
}
],
"container-title": "Life Sciences",
"container-title-short": "Life Sciences",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"elsevier.com",
"sciencedirect.com"
]
},
"created": {
"date-parts": [
[
2020,
4,
4
]
],
"date-time": "2020-04-04T00:26:44Z",
"timestamp": 1585960004000
},
"deposited": {
"date-parts": [
[
2020,
4,
22
]
],
"date-time": "2020-04-22T08:02:01Z",
"timestamp": 1587542521000
},
"funder": [
{
"DOI": "10.13039/501100004686",
"award": [
"1811016"
],
"doi-asserted-by": "publisher",
"name": "Deanship of Scientific Research, King Faisal University"
}
],
"indexed": {
"date-parts": [
[
2023,
9,
28
]
],
"date-time": "2023-09-28T17:04:12Z",
"timestamp": 1695920652201
},
"is-referenced-by-count": 260,
"issued": {
"date-parts": [
[
2020,
6
]
]
},
"language": "en",
"license": [
{
"URL": "https://www.elsevier.com/tdm/userlicense/1.0/",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2020,
6,
1
]
],
"date-time": "2020-06-01T00:00:00Z",
"timestamp": 1590969600000
}
}
],
"link": [
{
"URL": "https://api.elsevier.com/content/article/PII:S0024320520303751?httpAccept=text/xml",
"content-type": "text/xml",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://api.elsevier.com/content/article/PII:S0024320520303751?httpAccept=text/plain",
"content-type": "text/plain",
"content-version": "vor",
"intended-application": "text-mining"
}
],
"member": "78",
"original-title": [],
"page": "117627",
"prefix": "10.1016",
"published": {
"date-parts": [
[
2020,
6
]
]
},
"published-print": {
"date-parts": [
[
2020,
6
]
]
},
"publisher": "Elsevier BV",
"reference": [
{
"article-title": "Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission",
"author": "Xu",
"first-page": "1",
"journal-title": "Sci. China Life Sci.",
"key": "10.1016/j.lfs.2020.117627_bb0005",
"year": "2020"
},
{
"DOI": "10.1016/S0140-6736(03)13077-2",
"article-title": "Coronavirus as a possible cause of severe acute respiratory syndrome",
"author": "Peiris",
"doi-asserted-by": "crossref",
"first-page": "1319",
"journal-title": "Lancet",
"key": "10.1016/j.lfs.2020.117627_bb0010",
"volume": "361",
"year": "2003"
},
{
"DOI": "10.1128/JVI.80.9.4211-4219.2006",
"article-title": "Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "4211",
"journal-title": "J. Virol.",
"key": "10.1016/j.lfs.2020.117627_bb0015",
"volume": "80",
"year": "2006"
},
{
"DOI": "10.1056/NEJMoa1211721",
"article-title": "Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia",
"author": "Zaki",
"doi-asserted-by": "crossref",
"first-page": "1814",
"journal-title": "N. Engl. J. Med.",
"key": "10.1016/j.lfs.2020.117627_bb0020",
"volume": "367",
"year": "2012"
},
{
"DOI": "10.3201/eid1002.030351",
"article-title": "Introduction of SARS in France, March–April, 2003",
"author": "Desenclos",
"doi-asserted-by": "crossref",
"first-page": "195",
"journal-title": "Emerg. Infect. Dis.",
"key": "10.1016/j.lfs.2020.117627_bb0025",
"volume": "10",
"year": "2004"
},
{
"DOI": "10.1093/ajcp/aqaa029",
"article-title": "Three emerging coronaviruses in two decades: the story of SARS, MERS, and now COVID-19",
"author": "Guarner",
"doi-asserted-by": "crossref",
"first-page": "420",
"issue": "4",
"journal-title": "Am. J. Clin. Pathol.",
"key": "10.1016/j.lfs.2020.117627_bb0030",
"volume": "153",
"year": "2020"
},
{
"DOI": "10.1128/JVI.00127-20",
"article-title": "Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS",
"author": "Wan",
"doi-asserted-by": "crossref",
"first-page": "e00127",
"issue": "7",
"journal-title": "J. Virol.",
"key": "10.1016/j.lfs.2020.117627_bb0035",
"volume": "94",
"year": "2020"
},
{
"DOI": "10.1111/febs.12936",
"article-title": "From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design",
"author": "Hilgenfeld",
"doi-asserted-by": "crossref",
"first-page": "4085",
"journal-title": "FEBS J.",
"key": "10.1016/j.lfs.2020.117627_bb0040",
"volume": "281",
"year": "2014"
},
{
"DOI": "10.2174/1570180815666180918161922",
"article-title": "Molecular dynamics and inhibition of MERS CoV papain-like protease by small molecule imidazole and aminopurine derivatives",
"author": "Kandeel",
"doi-asserted-by": "crossref",
"first-page": "584",
"journal-title": "Lett. Drug Des. Discov.",
"key": "10.1016/j.lfs.2020.117627_bb0045",
"volume": "16",
"year": "2019"
},
{
"DOI": "10.1016/j.eng.2018.11.035",
"article-title": "Molecular characteristics, functions, and related pathogenicity of MERS-CoV proteins",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "940",
"issue": "5",
"journal-title": "Engineering",
"key": "10.1016/j.lfs.2020.117627_bb0050",
"volume": "5",
"year": "2019"
},
{
"DOI": "10.1016/j.drudis.2020.01.015",
"article-title": "Recent discovery and development of inhibitors targeting coronaviruses",
"author": "Pillaiyar",
"doi-asserted-by": "crossref",
"journal-title": "Drug Discov. Today",
"key": "10.1016/j.lfs.2020.117627_bb0055",
"year": "2020"
},
{
"DOI": "10.1038/nrd.2015.37",
"article-title": "Coronaviruses—drug discovery and therapeutic options",
"author": "Zumla",
"doi-asserted-by": "crossref",
"first-page": "327",
"journal-title": "Nat. Rev. Drug Discov.",
"key": "10.1016/j.lfs.2020.117627_bb0060",
"volume": "15",
"year": "2016"
},
{
"DOI": "10.1021/jm070295s",
"article-title": "Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus",
"author": "Wen",
"doi-asserted-by": "crossref",
"first-page": "4087",
"journal-title": "J. Med. Chem.",
"key": "10.1016/j.lfs.2020.117627_bb0065",
"volume": "50",
"year": "2007"
},
{
"DOI": "10.1080/07391102.2015.1014423",
"article-title": "Molecular analysis of some camel cytochrome P450 enzymes reveals lower evolution and drug-binding properties",
"author": "Altaher",
"doi-asserted-by": "crossref",
"first-page": "115",
"journal-title": "J. Biomol. Struct. Dyn.",
"key": "10.1016/j.lfs.2020.117627_bb0070",
"volume": "34",
"year": "2016"
},
{
"DOI": "10.3923/ijp.2015.243.247",
"article-title": "Annotation of camel genome for estimation of drug binding power, evolution and adaption of cytochrome P450 1a2",
"author": "Altaher",
"doi-asserted-by": "crossref",
"first-page": "243",
"journal-title": "Int. J. Pharmacol.",
"key": "10.1016/j.lfs.2020.117627_bb0075",
"volume": "11",
"year": "2015"
},
{
"DOI": "10.1017/S0031182008005301",
"article-title": "Mutational, inhibitory and microcalorimetric analyses of Plasmodium falciparum TMP kinase. Implications for drug discovery",
"author": "Kandeel",
"doi-asserted-by": "crossref",
"first-page": "11",
"journal-title": "Parasitology",
"key": "10.1016/j.lfs.2020.117627_bb0080",
"volume": "136",
"year": "2009"
},
{
"DOI": "10.1016/j.ddtec.2012.07.009",
"article-title": "In silico virtual screening approaches for anti-viral drug discovery",
"author": "Murgueitio",
"doi-asserted-by": "crossref",
"journal-title": "Drug Discov. Today Technol.",
"key": "10.1016/j.lfs.2020.117627_bb0085",
"volume": "9",
"year": "2012"
},
{
"DOI": "10.1016/j.jviromet.2019.113806",
"article-title": "Analysis of preferred codon usage in the coronavirus N genes and their implications for genome evolution and vaccine design",
"author": "Sheikh",
"doi-asserted-by": "crossref",
"journal-title": "J. Virol. Methods",
"key": "10.1016/j.lfs.2020.117627_bb0090",
"volume": "277",
"year": "2020"
},
{
"DOI": "10.1002/jcc.20186",
"article-title": "SARS-CoV protease inhibitors design using virtual screening method from natural products libraries",
"author": "Liu",
"doi-asserted-by": "crossref",
"first-page": "484",
"journal-title": "J. Comput. Chem.",
"key": "10.1016/j.lfs.2020.117627_bb0095",
"volume": "26",
"year": "2005"
},
{
"DOI": "10.1021/ci034270n",
"article-title": "Virtual screening for SARS-CoV protease based on KZ7088 pharmacophore points",
"author": "Sirois",
"doi-asserted-by": "crossref",
"first-page": "1111",
"journal-title": "J. Chem. Inf. Comput. Sci.",
"key": "10.1016/j.lfs.2020.117627_bb0100",
"volume": "44",
"year": "2004"
},
{
"DOI": "10.1186/1471-2105-13-S17-S5",
"article-title": "Multiple virtual screening approaches for finding new Hepatitis c virus RNA-dependent RNA polymerase inhibitors: structure-based screens and molecular dynamics for the pursue of new poly pharmacological inhibitors",
"author": "ElHefnawi",
"doi-asserted-by": "crossref",
"first-page": "S5",
"journal-title": "BMC Bioinf. BioMed Central",
"key": "10.1016/j.lfs.2020.117627_bb0105",
"year": "2012"
},
{
"DOI": "10.1021/cb800176t",
"article-title": "Antiviral compounds discovered by virtual screening of small− molecule libraries against dengue virus E protein",
"author": "Zhou",
"doi-asserted-by": "crossref",
"first-page": "765",
"journal-title": "ACS Chem. Biol.",
"key": "10.1016/j.lfs.2020.117627_bb0110",
"volume": "3",
"year": "2008"
},
{
"DOI": "10.1007/s12539-015-0109-8",
"article-title": "Flavonoids as multi-target inhibitors for proteins associated with Ebola virus: in silico discovery using virtual screening and molecular docking studies",
"author": "Raj",
"doi-asserted-by": "crossref",
"first-page": "132",
"journal-title": "Interdiscip. Sci. Comput. Life Sci.",
"key": "10.1016/j.lfs.2020.117627_bb0115",
"volume": "8",
"year": "2016"
},
{
"DOI": "10.1038/82191",
"article-title": "The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen",
"author": "Crotty",
"doi-asserted-by": "crossref",
"first-page": "1375",
"journal-title": "Nat. Med.",
"key": "10.1016/j.lfs.2020.117627_bb0120",
"volume": "6",
"year": "2000"
},
{
"article-title": "Ribavirin in the treatment of SARS: a new trick for an old drug?",
"author": "Koren",
"first-page": "1289",
"journal-title": "CMAJ",
"key": "10.1016/j.lfs.2020.117627_bb0125",
"volume": "168",
"year": "2003"
},
{
"DOI": "10.2165/11204330-000000000-00000",
"article-title": "Telbivudine",
"author": "McKeage",
"doi-asserted-by": "crossref",
"first-page": "1857",
"journal-title": "Drugs",
"key": "10.1016/j.lfs.2020.117627_bb0130",
"volume": "70",
"year": "2010"
},
{
"DOI": "10.1021/jm0306430",
"article-title": "Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy",
"author": "Friesner",
"doi-asserted-by": "crossref",
"first-page": "1739",
"issue": "7",
"journal-title": "J. Med. Chem.",
"key": "10.1016/j.lfs.2020.117627_bb9000",
"volume": "47",
"year": "2004"
},
{
"DOI": "10.1021/jm030644s",
"article-title": "Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening",
"author": "Halgren",
"doi-asserted-by": "crossref",
"first-page": "1750",
"issue": "7",
"journal-title": "J. Med. Chem.",
"key": "10.1016/j.lfs.2020.117627_bb8000",
"volume": "47",
"year": "2004"
}
],
"reference-count": 28,
"references-count": 28,
"relation": {},
"resource": {
"primary": {
"URL": "https://linkinghub.elsevier.com/retrieve/pii/S0024320520303751"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"General Pharmacology, Toxicology and Pharmaceutics",
"General Biochemistry, Genetics and Molecular Biology",
"General Medicine"
],
"subtitle": [],
"title": "Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1016/elsevier_cm_policy",
"volume": "251"
}