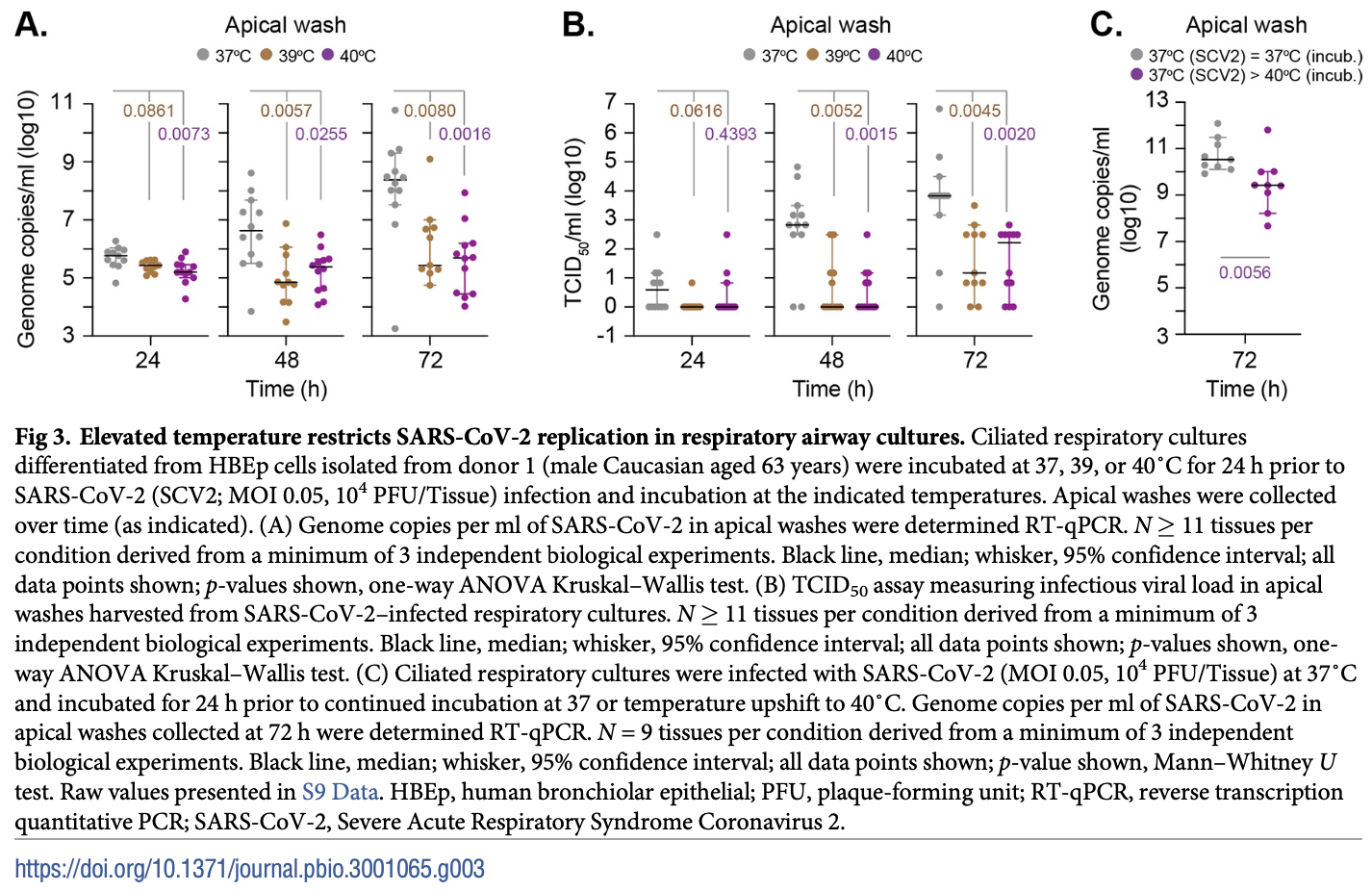
Elevated temperature inhibits SARS-CoV-2 replication in respiratory epithelium independently of IFN-mediated innate immune defenses
et al., PLOS Biology, doi:10.1371/journal.pbio.3001065, Dec 2021
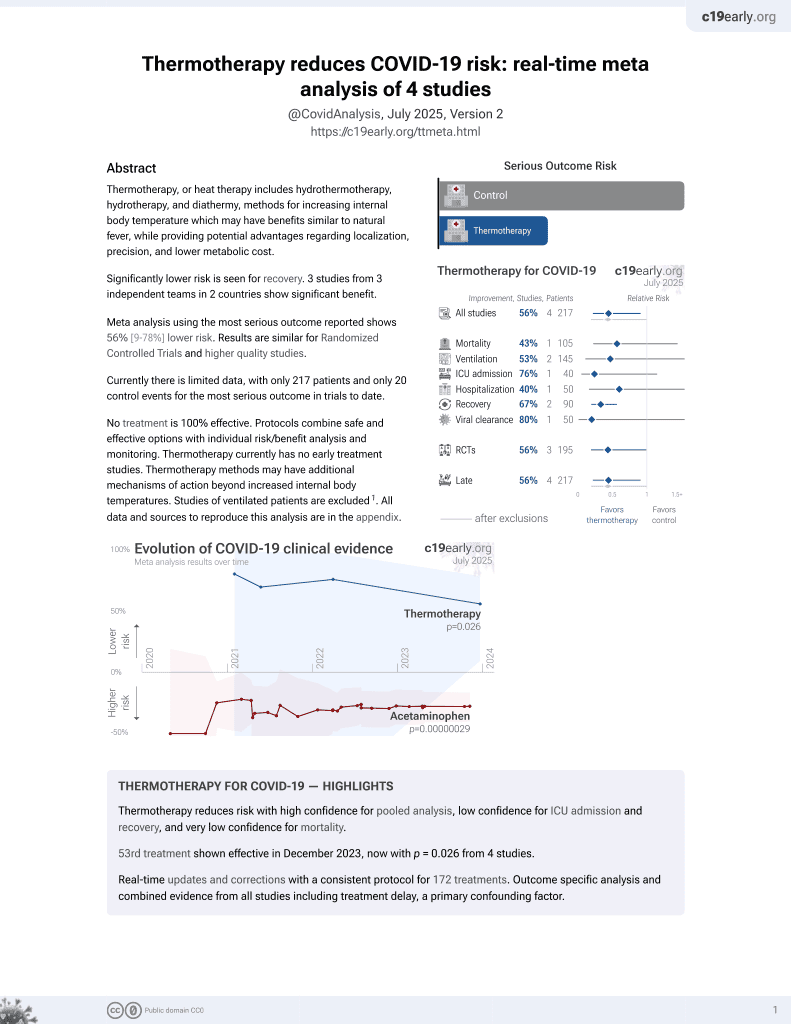
54th treatment shown to reduce risk in
December 2023, now with p = 0.026 from 4 studies.
Lower risk for recovery.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
In vitro study using a 3D respiratory epithelial model and cells from human donors, showing that elevated temperature (39-40°C) restricts SARS-CoV-2 infection and replication independently of interferon-mediated antiviral defenses. Authors found SARS-CoV-2 can still enter respiratory cells at 40°C but viral transcription and replication are inhibited, limiting infectious virus production. This temperature-dependent restriction correlates with altered host gene expression related to antiviral immunity and epigenetic regulation. As fever is a common COVID-19 symptom, the data suggests febrile temperature ranges may confer protection to respiratory tissues by restricting SARS-CoV-2 propagation.
Herder et al., 21 Dec 2021, United Kingdom, peer-reviewed, 22 authors.
Contact: sheila.graham@glasgow.ac.uk, pablo.murcia@glasgow.ac.uk, chris.boutell@glasgow.ac.uk.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Elevated temperature inhibits SARS-CoV-2 replication in respiratory epithelium independently of IFN-mediated innate immune defenses
PLOS Biology, doi:10.1371/journal.pbio.3001065
AU : Pleaseconfirmthatallheadinglevelsarerepresentedcorrectly: The pandemic spread of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), the etiological agent of Coronavirus Disease 2019 (COVID-19), represents an ongoing international health crisis. A key symptom of SARS-CoV-2 infection is the onset of fever, with a hyperthermic temperature range of 38 to 41˚C. Fever is an evolutionarily conserved host response to microbial infection that can influence the outcome of viral pathogenicity and regulation of host innate and adaptive immune responses. However, it remains to be determined what effect elevated temperature has on SARS-CoV-2 replication. Utilizing a three-dimensional (3D) air-liquid interface (ALI) model that closely mimics the natural tissue physiology of SARS-CoV-2 infection in the respiratory airway, we identify tissue temperature to play an important role in the regulation of SARS-CoV-2 infection. Respiratory tissue incubated at 40˚C remained permissive to SARS-CoV-2 entry but refractory to viral transcription, leading to significantly reduced levels of viral RNA replication and apical shedding of infectious virus. We identify tissue temperature to play an important role in the differential regulation of epithelial host responses to SARS-CoV-2 infection that impact upon multiple pathways, including intracellular immune regulation, without disruption to general transcription or epithelium integrity. We present the first evidence that febrile temperatures associated with COVID-19 inhibit SARS-CoV-2 replication in respiratory epithelia. Our data identify an important role for tissue temperature in the epithelial restriction of SARS-CoV-2 independently of canonical interferon (IFN)-mediated antiviral immune defenses.
Author Contributions Conceptualization: Vanessa Herder, Kieran Dee, Ruth F. Jarrett, Sheila V. Graham, Pablo R. Murcia, Chris Boutell.
References
Blanco-Melo, Nilsson-Payant, Liu, Uhl, Hoagland et al., Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19, Cell, doi:10.1016/j.cell.2020.04.026
Bradley, Maioli, Johnston, Chaudhry, Fink et al., Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series, Lancet, doi:10.1016/S0140-6736%2820%2931305-2
Cao, Li, Liang, Wang, Wei et al., Clinical characteristics of Coronavirus Disease 2019 patients in Beijing, China, PLoS ONE, doi:10.1371/journal.pone.0234764
Christianson, Ingersoll, Landon, Pfeiffer, Gerber, Characterization of a temperature sensitive feline infectious peritonitis coronavirus, Arch Virol, doi:10.1007/BF01311080
Chua, Lukassen, Trump, Hennig, Wendisch et al., COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis, Nat Biotechnol, doi:10.1038/s41587-020-0602-4
Curina, Termanini, Barozzi, Prosperini, Simonatto et al., High constitutive activity of a broad panel of housekeeping and tissue-specific cis-regulatory elements depends on a subset of ETS proteins, Genes Dev, doi:10.1101/gad.293134.116
Dalton, Mullin, Amorim, Medcalf, Tiley et al., Temperature sensitive influenza A virus genome replication results from low thermal stability of polymerase-cRNA complexes, Virol J, doi:10.1186/1743-422X-3-58
Deng, Mettelman, 'brien, Thompson, Brien, Analysis of Coronavirus Temperature-Sensitive Mutants Reveals an Interplay between the Macrodomain and Papain-Like Protease Impacting Replication and Pathogenesis, J Virol, doi:10.1128/JVI.02140-18
Deshmukh, Motwani, Kumar, Kumari, Raza, Histopathological observations in COVID-19: a systematic review, J Clin Pathol, doi:10.1136/jclinpath-2020-206995
Desmyter, Melnick, Rawls, Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero), J Virol, doi:10.1128/JVI.2.10.955-961.1968
Earn, Andrews, Bolker, Population-level effects of suppressing fever, Proc Biol Sci, doi:10.1098/rspb.2013.2570
Eisenberg, Levanon, Human housekeeping genes, revisited, Trends Genet, doi:10.1016/j.tig.2013.05.010
Evans, Repasky, Fisher, Fever and the thermal regulation of immunity: the immune system feels the heat, Nat Rev Immunol, doi:10.1038/nri3843
Fabregat, Jupe, Matthews, Sidiropoulos, Gillespie et al., The Reactome Pathway Knowledgebase, Nucleic Acids Res, doi:10.1093/nar/gkx1132
Felgenhauer, Schoen, Gad, Hartmann, Schaubmar et al., Inhibition of SARS-CoV-2 by type I and type III interferons, J Biol Chem, doi:10.1074/jbc.AC120.013788
Flohr, Schneider-Schaulies, Haller, Kochs, The central interactive region of human MxA GTPase is involved in GTPase activation and interaction with viral target structures, FEBS Lett, doi:10.1016/s0014-5793%2899%2901598-7
Foxman, Storer, Fitzgerald, Wasik, Hou et al., Temperature-dependent innate defense against the common cold virus limits viral replication at warm temperature in mouse airway cells, Proc Natl Acad Sci U S A, doi:10.1073/pnas.1411030112
Fu, Wang, Yuan, Chen, Ao et al., Clinical characteristics of coronavirus disease 2019 (COVID-19) in China: A systematic review and meta-analysis, J Infect, doi:10.1016/j.jinf.2020.03.041
Grubaugh, Gangavarapu, Quick, Matteson, Jesus et al., An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar, Genome Biol, doi:10.1186/s13059-018-1618-7
Guan, Ni, Hu, Liang, Ou et al., Clinical Characteristics of Coronavirus Disease 2019 in China, N Engl J Med, doi:10.1056/NEJMoa2002032
Hadjadj, Yatim, Barnabei, Corneau, Boussier et al., Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients, Science, doi:10.1126/science.abc6027
Hao, Ning, Kuz, Vorhies, Yan et al., Long-Term Modeling of SARS-CoV-2 Infection of In Vitro Cultured Polarized Human Airway Epithelium, mBio, doi:10.1128/mBio.02852-20
Hassan, Case, Winkler, Thackray, Kafai et al., A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies, Cell, doi:10.1016/j.cell.2020.06.011
Hierholzer, Killington, 2-Virus isolation and quantitation, Virology Methods Manual
Hirayama, Atagi, Hiraki, Kim, Heat shock protein 70 is related to thermal inhibition of nuclear export of the influenza virus ribonucleoprotein complex, J Virol, doi:10.1128/jvi.78.3.1263-1270.2004
Hoang, Chorath, Moreira, Evans, Burmeister-Morton et al., COVID-19 in 7780 pediatric patients: A systematic review, EClinicalMedicine, doi:10.1016/j.eclinm.2020.100433
Hoffmann, Kleine-Weber, Schroeder, Kruger, Herrler et al., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor, Cell, doi:10.1016/j.cell.2020.02.052
Huang, Wang, Li, Ren, Zhao et al., Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, Lancet, doi:10.1016/S0140-6736%2820%2930183-5
Jassal, Matthews, Viteri, Gong, Lorente et al., The reactome pathway knowledgebase, Nucleic Acids Res, doi:10.1093/nar/gkz1031
Killip, Jackson, Perez-Cidoncha, Fodor, Randall, Single-cell studies of IFN-beta promoter activation by wild-type and NS1-defective influenza A viruses, J Gen Virol, doi:10.1099/jgv.0.000687
Kim, Kim, Kim, Kim, Park et al., Infection and Rapid Transmission of SARS-CoV-2 in Ferrets, Cell Host Microbe, doi:10.1016/j.chom.2020.03.023
Kim, Langmead, Salzberg, HISAT: a fast spliced aligner with low memory requirements, Nat Methods, doi:10.1038/nmeth.3317
Kim, Lee, Yang, Kim, Kim et al., The Architecture of SARS-CoV-2 Transcriptome, Cell, doi:10.1016/j.cell.2020.04.011
Kindler, Jonsdottir, Muth, Hamming, Hartmann et al., Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential, mBio, doi:10.1128/mBio.00611-12
Li, Durbin, Fast and accurate short read alignment with Burrows-Wheeler transform, Bioinformatics, doi:10.1093/bioinformatics/btp324
Li, Moore, Vasilieva, Sui, Wong et al., Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus, Nature, doi:10.1038/nature02145
Li, Zhang, Tong, Liu, Ye, Heat shock protein 70 inhibits the activity of Influenza A virus ribonucleoprotein and blocks the replication of virus in vitro and in vivo, PLoS ONE, doi:10.1371/journal.pone.0016546
Liao, Smyth, Shi, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features, Bioinformatics, doi:10.1093/bioinformatics/btt656
Long, Mistry, Haslam, Barclay, Host and viral determinants of influenza A virus species specificity, Nat Rev Microbiol, doi:10.1038/s41579-018-0115-z
Lu, Zhao, Li, Niu, Yang et al., Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding, Lancet, doi:10.1016/S0140-6736%2820%2930251-8
Milewska, Kula-Pacurar, Wadas, Suder, Szczepanski et al., Replication of Severe Acute Respiratory Syndrome Coronavirus 2 in Human Respiratory Epithelium, J Virol, doi:10.1128/JVI.00957-20
Moein, Javanmard, Abedi, Izadpanahi, Gheisari, Identification of Appropriate Housekeeping Genes for Gene Expression Analysis in Long-term Hypoxia-treated Kidney Cells, Adv Biomed Res, doi:10.4103/2277-9175.200790
Morimoto, Cells in stress: transcriptional activation of heat shock genes, Science, doi:10.1126/science.8451637
Mosca, Pitha, Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis, Mol Cell Biol, doi:10.1128/mcb.6.6.2279-2283.1986
Muralidharan, Mandrekar, Cellular stress response and innate immune signaling: integrating pathways in host defense and inflammation, J Leukoc Biol, doi:10.1189/jlb.0313153
Neubauer, Johow, Mack, Burchert, Meyn et al., The janus-kinase inhibitor ruxolitinib in SARS-CoV-2 induced acute respiratory distress syndrome (ARDS), Leukemia, doi:10.1038/s41375-021-01374-3
Ogoina, Fever, fever patterns and diseases called 'fever'-a review, J Infect Public Health, doi:10.1016/j.jiph.2011.05.002
Oyler-Yaniv, Oyler-Yaniv, Shakiba, Min, Chen et al., Catch and Release of Cytokines Mediated by Tumor Phosphatidylserine Converts Transient Exposure into Long-Lived Inflammation, Mol Cell, doi:10.1016/j.molcel.2017.05.011
Oyler-Yaniv, Oyler-Yaniv, Whitlock, Liu, Germain et al., A Tunable Diffusion-Consumption Mechanism of Cytokine Propagation Enables Plasticity in Cell-to-Cell Communication in the Immune System, Immunity, doi:10.1016/j.immuni.2017.03.011
Parker, Lindsey, Leary, Gaudieri, Chopra et al., Subgenomic RNA identification in SARS-CoV-2 genomic sequencing data, Genome Res, doi:10.1101/gr.268110.120
Pizzorno, Padey, Julien, Trouillet-Assant, Traversier et al., Characterization and Treatment of SARS-CoV-2 in Nasal and Bronchial Human Airway Epithelia, Cell Rep Med, doi:10.1016/j.xcrm.2020.100059
Polak, Van Gool, Cohen, Der Thusen, Van Paassen, A systematic review of pathological findings in COVID-19: a pathophysiological timeline and possible mechanisms of disease progression, Mod Pathol, doi:10.1038/s41379-020-0603-3
Ravindra, Alfajaro, Gasque, Huston, Wan et al., Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes, doi:10.1371/journal.pbio.3001143
Robinson, Mccarthy, Smyth, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics, doi:10.1093/bioinformatics/btp616
Russell, Elshina, Kowalsky, Velthuis, Bloom, Single-Cell Virus Sequencing of Influenza Infections That Trigger Innate Immunity, J Virol, doi:10.1128/JVI.00500-19
Ryan, Levy, Clinical review: fever in intensive care unit patients, Crit Care, doi:10.1186/cc1879
Schulman, Namias, Doherty, Manning, Li et al., The effect of antipyretic therapy upon outcomes in critically ill patients: a randomized, prospective study, Surg Infect, doi:10.1089/sur.2005.6.369
Sia, Yan, Chin, Fung, Choy et al., Pathogenesis and transmission of SARS-CoV-2 in golden hamsters, Nature, doi:10.1038/s41586-020-2342-5
Stanifer, Kee, Zumaran, Triana, Mukenhirn, Critical Role of Type III Interferon in Controlling SARS-CoV-2 Infection in Human Intestinal Epithelial Cells, Cell Rep, doi:10.1016/j.celrep.2020.107863
Stewart, Randall, Adamson, Inhibitors of the interferon response enhance virus replication in vitro, PLoS ONE, doi:10.1371/journal.pone.0112014
Tapia, Kim, Sun, Mercado-Lopez, Dunay et al., Defective viral genomes arising in vivo provide critical danger signals for the triggering of lung antiviral immunity, PLoS Pathog, doi:10.1371/journal.ppat.1003703
Triana, Metz-Zumaran, Ramirez, Kee, Doldan et al., Single-cell analyses reveal SARS-CoV-2 interference with intrinsic immune response in the human gut, Mol Syst Biol, doi:10.15252/msb.202110232
Trinklein, Murray, Hartman, Botstein, Myers, The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response, Mol Biol Cell, doi:10.1091/mbc.e03-10-0738
Trypsteen, Van Cleemput, Snippenberg, Gerlo, Vandekerckhove, On the whereabouts of SARS-CoV-2 in the human body: A systematic review, PLoS Pathog, doi:10.1371/journal.ppat.1009037
V'kovski, Gultom, Kelly, Steiner, Russeil et al., Disparate temperature-dependent virus-host dynamics for SARS-CoV-2 and SARS-CoV in the human respiratory epithelium, PLoS Biol, doi:10.1371/journal.pbio.3001158
Vasilijevic, Zamarreno, Oliveros, Rodriguez-Frandsen, Gomez et al., Reduced accumulation of defective viral genomes contributes to severe outcome in influenza virus infected patients, PLoS Pathog, doi:10.1371/journal.ppat.1006650
Wan, Song, Li, He, Stress proteins: the biological functions in virus infection, present and challenges for target-based antiviral drug development, Signal Transduct Target Ther, doi:10.1038/s41392-020-00233-4
Wang, Xu, Wang, Cao, An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism, Science, doi:10.1126/science.aao0409
Wu, Zhao, Yu, Chen, Song, Author Correction: A new coronavirus associated with human respiratory disease in China, Nature, doi:10.1038/s41586-020-2202-3
Xu, Chen, Zhu, Cui, Chen et al., Clinical features and dynamics of viral load in imported and non-imported patients with COVID-19, Int J Infect Dis, doi:10.1016/j.ijid.2020.03.022
Youk, Kim, Evans, Jeong, Hur et al., Three-Dimensional Human Alveolar Stem Cell Culture Models Reveal Infection Response to SARS-CoV-2, Cell Stem Cell, doi:10.1016/j.stem.2020.10.004
Zaeck, Scheibner, Sehl, Muller, Hoffmann et al., Light Sheet Microscopy-Assisted 3D Analysis of SARS-CoV-2 Infection in the Respiratory Tract of the Ferret Model, Viruses, doi:10.3390/v13030529
Zhou, Zhou, Pache, Chang, Khodabakhshi et al., Metascape provides a biologist-oriented resource for the analysis of systems-level datasets, Nat Commun, doi:10.1038/s41467-019-09234-6
Zhu, Wang, Liu, Liang, Wang et al., Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells, Nat Commun, doi:10.1038/s41467-020-17796-z
Zhu, Zhang, Li, Yang, Song, A Novel Coronavirus from Patients with Pneumonia in China, N Engl J Med, doi:10.1056/NEJMoa2001017
Ziegler, Allon, Nyquist, Mbano, Miao et al., SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues, Cell, doi:10.1016/j.cell.2020.04.035
DOI record:
{
"DOI": "10.1371/journal.pbio.3001065",
"ISSN": [
"1545-7885"
],
"URL": "http://dx.doi.org/10.1371/journal.pbio.3001065",
"abstract": "<jats:p>The pandemic spread of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), the etiological agent of Coronavirus Disease 2019 (COVID-19), represents an ongoing international health crisis. A key symptom of SARS-CoV-2 infection is the onset of fever, with a hyperthermic temperature range of 38 to 41°C. Fever is an evolutionarily conserved host response to microbial infection that can influence the outcome of viral pathogenicity and regulation of host innate and adaptive immune responses. However, it remains to be determined what effect elevated temperature has on SARS-CoV-2 replication. Utilizing a three-dimensional (3D) air–liquid interface (ALI) model that closely mimics the natural tissue physiology of SARS-CoV-2 infection in the respiratory airway, we identify tissue temperature to play an important role in the regulation of SARS-CoV-2 infection. Respiratory tissue incubated at 40°C remained permissive to SARS-CoV-2 entry but refractory to viral transcription, leading to significantly reduced levels of viral RNA replication and apical shedding of infectious virus. We identify tissue temperature to play an important role in the differential regulation of epithelial host responses to SARS-CoV-2 infection that impact upon multiple pathways, including intracellular immune regulation, without disruption to general transcription or epithelium integrity. We present the first evidence that febrile temperatures associated with COVID-19 inhibit SARS-CoV-2 replication in respiratory epithelia. Our data identify an important role for tissue temperature in the epithelial restriction of SARS-CoV-2 independently of canonical interferon (IFN)-mediated antiviral immune defenses.</jats:p>",
"author": [
{
"ORCID": "http://orcid.org/0000-0003-4814-1382",
"affiliation": [],
"authenticated-orcid": true,
"family": "Herder",
"given": "Vanessa",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0002-3280-6768",
"affiliation": [],
"authenticated-orcid": true,
"family": "Dee",
"given": "Kieran",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-4345-3475",
"affiliation": [],
"authenticated-orcid": true,
"family": "Wojtus",
"given": "Joanna K.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-2056-0510",
"affiliation": [],
"authenticated-orcid": true,
"family": "Epifano",
"given": "Ilaria",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-6014-387X",
"affiliation": [],
"authenticated-orcid": true,
"family": "Goldfarb",
"given": "Daniel",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-2438-5219",
"affiliation": [],
"authenticated-orcid": true,
"family": "Rozario",
"given": "Christoforos",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-1201-6734",
"affiliation": [],
"authenticated-orcid": true,
"family": "Gu",
"given": "Quan",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-9442-2903",
"affiliation": [],
"authenticated-orcid": true,
"family": "Da Silva Filipe",
"given": "Ana",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Nomikou",
"given": "Kyriaki",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-0093-7170",
"affiliation": [],
"authenticated-orcid": true,
"family": "Nichols",
"given": "Jenna",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Jarrett",
"given": "Ruth F.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5325-5360",
"affiliation": [],
"authenticated-orcid": true,
"family": "Stevenson",
"given": "Andrew",
"sequence": "additional"
},
{
"affiliation": [],
"family": "McFarlane",
"given": "Steven",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-6275-2740",
"affiliation": [],
"authenticated-orcid": true,
"family": "Stewart",
"given": "Meredith E.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-3085-9994",
"affiliation": [],
"authenticated-orcid": true,
"family": "Szemiel",
"given": "Agnieszka M.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-3291-1397",
"affiliation": [],
"authenticated-orcid": true,
"family": "Pinto",
"given": "Rute M.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-2094-3001",
"affiliation": [],
"authenticated-orcid": true,
"family": "Masdefiol Garriga",
"given": "Andreu",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-7317-3266",
"affiliation": [],
"authenticated-orcid": true,
"family": "Davis",
"given": "Chris",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-6482-7125",
"affiliation": [],
"authenticated-orcid": true,
"family": "Allan",
"given": "Jay",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-7140-8279",
"affiliation": [],
"authenticated-orcid": true,
"family": "Graham",
"given": "Sheila V.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-4352-394X",
"affiliation": [],
"authenticated-orcid": true,
"family": "Murcia",
"given": "Pablo R.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-2970-7785",
"affiliation": [],
"authenticated-orcid": true,
"family": "Boutell",
"given": "Chris",
"sequence": "additional"
}
],
"container-title": "PLOS Biology",
"container-title-short": "PLoS Biol",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"www.plosbiology.org"
]
},
"created": {
"date-parts": [
[
2021,
12,
21
]
],
"date-time": "2021-12-21T18:50:58Z",
"timestamp": 1640112658000
},
"deposited": {
"date-parts": [
[
2022,
1,
18
]
],
"date-time": "2022-01-18T20:34:56Z",
"timestamp": 1642538096000
},
"editor": [
{
"affiliation": [],
"family": "Cimarelli",
"given": "Andrea",
"sequence": "first"
}
],
"funder": [
{
"DOI": "10.13039/501100001659",
"award": [
"406109949"
],
"doi-asserted-by": "publisher",
"name": "Deutsche Forschungsgemeinschaft"
},
{
"award": [
"01KI1723G"
],
"name": "BMEL Förderkennzeichen"
},
{
"DOI": "10.13039/501100000265",
"award": [
"MC_UU_12014/9"
],
"doi-asserted-by": "publisher",
"name": "Medical Research Council"
},
{
"DOI": "10.13039/501100000265",
"award": [
"MC_ST_U18018"
],
"doi-asserted-by": "publisher",
"name": "Medical Research Council"
},
{
"DOI": "10.13039/501100000265",
"award": [
"MR/R502327/1"
],
"doi-asserted-by": "publisher",
"name": "Medical Research Council"
},
{
"DOI": "10.13039/501100000268",
"award": [
"BB/R505341/1"
],
"doi-asserted-by": "publisher",
"name": "Biotechnology and Biological Sciences Research Council"
},
{
"DOI": "10.13039/501100000265",
"award": [
"MC_UU_12014/12"
],
"doi-asserted-by": "publisher",
"name": "Medical Research Council"
},
{
"DOI": "10.13039/501100000589",
"award": [
"TCS/19/11"
],
"doi-asserted-by": "publisher",
"name": "Chief Scientist Office"
},
{
"DOI": "10.13039/501100000265",
"award": [
"MC_PC_19026"
],
"doi-asserted-by": "publisher",
"name": "Medical Research Council"
},
{
"award": [
"BB/R019843/1"
],
"name": "UKRI/DHSC"
},
{
"DOI": "10.13039/501100000265",
"award": [
"MC_PC_19026"
],
"doi-asserted-by": "publisher",
"name": "Medical Research Council"
},
{
"award": [
"145813"
],
"name": "University of Glasgow School of Veterinary Medicine"
},
{
"award": [
"123939"
],
"name": "University of Glasgow School of Veterinary Medicine"
},
{
"DOI": "10.13039/501100000265",
"award": [
"MC_UU_12014/5"
],
"doi-asserted-by": "publisher",
"name": "Medical Research Council"
},
{
"DOI": "10.13039/501100000265",
"award": [
"MC_UU_12014/10"
],
"doi-asserted-by": "publisher",
"name": "Medical Research Council"
},
{
"DOI": "10.13039/501100000265",
"award": [
"MC_UU_12018/12"
],
"doi-asserted-by": "publisher",
"name": "Medical Research Council"
}
],
"indexed": {
"date-parts": [
[
2024,
1,
13
]
],
"date-time": "2024-01-13T18:41:59Z",
"timestamp": 1705171319001
},
"is-referenced-by-count": 19,
"issue": "12",
"issued": {
"date-parts": [
[
2021,
12,
21
]
]
},
"journal-issue": {
"issue": "12",
"published-online": {
"date-parts": [
[
2021,
12,
21
]
]
}
},
"language": "en",
"license": [
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
12,
21
]
],
"date-time": "2021-12-21T00:00:00Z",
"timestamp": 1640044800000
}
}
],
"link": [
{
"URL": "https://dx.plos.org/10.1371/journal.pbio.3001065",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "340",
"original-title": [],
"page": "e3001065",
"prefix": "10.1371",
"published": {
"date-parts": [
[
2021,
12,
21
]
]
},
"published-online": {
"date-parts": [
[
2021,
12,
21
]
]
},
"publisher": "Public Library of Science (PLoS)",
"reference": [
{
"DOI": "10.1056/NEJMoa2001017",
"article-title": "A Novel Coronavirus from Patients with Pneumonia in China, 2019",
"author": "N Zhu",
"doi-asserted-by": "crossref",
"first-page": "727",
"issue": "8",
"journal-title": "N Engl J Med",
"key": "pbio.3001065.ref001",
"volume": "382",
"year": "2020"
},
{
"DOI": "10.1016/S0140-6736(20)30251-8",
"article-title": "Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding",
"author": "R Lu",
"doi-asserted-by": "crossref",
"first-page": "565",
"issue": "10224",
"journal-title": "Lancet",
"key": "pbio.3001065.ref002",
"volume": "395",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2202-3",
"article-title": "Author Correction: A new coronavirus associated with human respiratory disease in China",
"author": "F Wu",
"doi-asserted-by": "crossref",
"first-page": "E7",
"issue": "7803",
"journal-title": "Nature",
"key": "pbio.3001065.ref003",
"volume": "580",
"year": "2020"
},
{
"DOI": "10.1016/S0140-6736(20)30183-5",
"article-title": "Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China.",
"author": "C Huang",
"doi-asserted-by": "crossref",
"first-page": "497",
"issue": "10223",
"journal-title": "Lancet",
"key": "pbio.3001065.ref004",
"volume": "395",
"year": "2020"
},
{
"DOI": "10.1056/NEJMoa2002032",
"article-title": "Clinical Characteristics of Coronavirus Disease 2019 in China",
"author": "WJ Guan",
"doi-asserted-by": "crossref",
"first-page": "1708",
"issue": "18",
"journal-title": "N Engl J Med",
"key": "pbio.3001065.ref005",
"volume": "382",
"year": "2020"
},
{
"DOI": "10.1016/j.jinf.2020.03.041",
"article-title": "Clinical characteristics of coronavirus disease 2019 (COVID-19) in China: A systematic review and meta-analysis.",
"author": "L Fu",
"doi-asserted-by": "crossref",
"first-page": "656",
"issue": "6",
"journal-title": "J Infect",
"key": "pbio.3001065.ref006",
"volume": "80",
"year": "2020"
},
{
"DOI": "10.1016/j.eclinm.2020.100433",
"article-title": "COVID-19 in 7780 pediatric patients: A systematic review.",
"author": "A Hoang",
"doi-asserted-by": "crossref",
"first-page": "100433",
"journal-title": "EClinicalMedicine",
"key": "pbio.3001065.ref007",
"volume": "24",
"year": "2020"
},
{
"DOI": "10.1016/j.ijid.2020.03.022",
"article-title": "Clinical features and dynamics of viral load in imported and non-imported patients with COVID-19",
"author": "T Xu",
"doi-asserted-by": "crossref",
"first-page": "68",
"journal-title": "Int J Infect Dis",
"key": "pbio.3001065.ref008",
"volume": "94",
"year": "2020"
},
{
"DOI": "10.1038/nri3843",
"article-title": "Fever and the thermal regulation of immunity: the immune system feels the heat",
"author": "SS Evans",
"doi-asserted-by": "crossref",
"first-page": "335",
"issue": "6",
"journal-title": "Nat Rev Immunol",
"key": "pbio.3001065.ref009",
"volume": "15",
"year": "2015"
},
{
"DOI": "10.1016/j.jiph.2011.05.002",
"article-title": "Fever, fever patterns and diseases called ’fever’—a review.",
"author": "D Ogoina",
"doi-asserted-by": "crossref",
"first-page": "108",
"issue": "3",
"journal-title": "J Infect Public Health",
"key": "pbio.3001065.ref010",
"volume": "4",
"year": "2011"
},
{
"DOI": "10.1073/pnas.1411030112",
"article-title": "Temperature-dependent innate defense against the common cold virus limits viral replication at warm temperature in mouse airway cells",
"author": "EF Foxman",
"doi-asserted-by": "crossref",
"first-page": "827",
"issue": "3",
"journal-title": "Proc Natl Acad Sci U S A",
"key": "pbio.3001065.ref011",
"volume": "112",
"year": "2015"
},
{
"article-title": "Population-level effects of suppressing fever",
"author": "DJ Earn",
"first-page": "20132570",
"issue": "1778",
"journal-title": "Proc Biol Sci",
"key": "pbio.3001065.ref012",
"volume": "281",
"year": "2014"
},
{
"DOI": "10.1089/sur.2005.6.369",
"article-title": "The effect of antipyretic therapy upon outcomes in critically ill patients: a randomized, prospective study.",
"author": "CI Schulman",
"doi-asserted-by": "crossref",
"first-page": "369",
"issue": "4",
"journal-title": "Surg Infect",
"key": "pbio.3001065.ref013",
"volume": "6",
"year": "2005"
},
{
"DOI": "10.1186/cc1879",
"article-title": "Clinical review: fever in intensive care unit patients.",
"author": "M Ryan",
"doi-asserted-by": "crossref",
"first-page": "221",
"issue": "3",
"journal-title": "Crit Care",
"key": "pbio.3001065.ref014",
"volume": "7",
"year": "2003"
},
{
"DOI": "10.1371/journal.pone.0234764",
"article-title": "Clinical characteristics of Coronavirus Disease 2019 patients in Beijing, China.",
"author": "Z Cao",
"doi-asserted-by": "crossref",
"first-page": "e0234764",
"issue": "6",
"journal-title": "PLoS ONE",
"key": "pbio.3001065.ref015",
"volume": "15",
"year": "2020"
},
{
"DOI": "10.1371/journal.pbio.3001158",
"article-title": "Disparate temperature-dependent virus-host dynamics for SARS-CoV-2 and SARS-CoV in the human respiratory epithelium",
"author": "P V’Kovski",
"doi-asserted-by": "crossref",
"first-page": "e3001158",
"issue": "3",
"journal-title": "PLoS Biol",
"key": "pbio.3001065.ref016",
"volume": "19",
"year": "2021"
},
{
"DOI": "10.1371/journal.pbio.3001143",
"article-title": "Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes.",
"author": "NG Ravindra",
"doi-asserted-by": "crossref",
"first-page": "e3001143",
"issue": "3",
"journal-title": "PLoS Biol",
"key": "pbio.3001065.ref017",
"volume": "19",
"year": "2021"
},
{
"DOI": "10.1038/s41467-020-17796-z",
"article-title": "Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells.",
"author": "N Zhu",
"doi-asserted-by": "crossref",
"first-page": "3910",
"issue": "1",
"journal-title": "Nat Commun",
"key": "pbio.3001065.ref018",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1038/s41587-020-0602-4",
"article-title": "COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis",
"author": "RL Chua",
"doi-asserted-by": "crossref",
"first-page": "970",
"issue": "8",
"journal-title": "Nat Biotechnol",
"key": "pbio.3001065.ref019",
"volume": "38",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.04.026",
"article-title": "Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19",
"author": "D Blanco-Melo",
"doi-asserted-by": "crossref",
"first-page": "1036",
"issue": "5",
"journal-title": "Cell",
"key": "pbio.3001065.ref020",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1016/j.xcrm.2020.100059",
"article-title": "Characterization and Treatment of SARS-CoV-2 in Nasal and Bronchial Human Airway Epithelia",
"author": "A Pizzorno",
"doi-asserted-by": "crossref",
"first-page": "100059",
"issue": "4",
"journal-title": "Cell Rep Med",
"key": "pbio.3001065.ref021",
"volume": "1",
"year": "2020"
},
{
"DOI": "10.1136/jclinpath-2020-206995",
"article-title": "Histopathological observations in COVID-19: a systematic review",
"author": "V Deshmukh",
"doi-asserted-by": "crossref",
"first-page": "76",
"issue": "2",
"journal-title": "J Clin Pathol",
"key": "pbio.3001065.ref022",
"volume": "74",
"year": "2021"
},
{
"article-title": "A systematic review of pathological findings in COVID-19: a pathophysiological timeline and possible mechanisms of disease progression.",
"author": "SB Polak",
"first-page": "2128",
"issue": "11",
"journal-title": "Mod Pathol.Epub 2020 Jun 24",
"key": "pbio.3001065.ref023",
"volume": "33",
"year": "2020"
},
{
"DOI": "10.1016/S0140-6736(20)31305-2",
"article-title": "Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series",
"author": "BT Bradley",
"doi-asserted-by": "crossref",
"first-page": "320",
"issue": "10247",
"journal-title": "Lancet",
"key": "pbio.3001065.ref024",
"volume": "396",
"year": "2020"
},
{
"DOI": "10.1038/nature02145",
"article-title": "Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus",
"author": "W Li",
"doi-asserted-by": "crossref",
"first-page": "450",
"issue": "6965",
"journal-title": "Nature",
"key": "pbio.3001065.ref025",
"volume": "426",
"year": "2003"
},
{
"DOI": "10.1016/j.cell.2020.02.052",
"article-title": "SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor",
"author": "M Hoffmann",
"doi-asserted-by": "crossref",
"first-page": "271",
"issue": "2",
"journal-title": "Cell",
"key": "pbio.3001065.ref026",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.04.035",
"article-title": "SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues",
"author": "CGK Ziegler",
"doi-asserted-by": "crossref",
"first-page": "1016",
"issue": "5",
"journal-title": "Cell",
"key": "pbio.3001065.ref027",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1371/journal.ppat.1009037",
"article-title": "On the whereabouts of SARS-CoV-2 in the human body: A systematic review.",
"author": "W Trypsteen",
"doi-asserted-by": "crossref",
"first-page": "e1009037",
"issue": "10",
"journal-title": "PLoS Pathog.",
"key": "pbio.3001065.ref028",
"volume": "16",
"year": "2020"
},
{
"DOI": "10.3390/v13030529",
"article-title": "Light Sheet Microscopy-Assisted 3D Analysis of SARS-CoV-2 Infection in the Respiratory Tract of the Ferret Model.",
"author": "LM Zaeck",
"doi-asserted-by": "crossref",
"issue": "3",
"journal-title": "Viruses",
"key": "pbio.3001065.ref029",
"volume": "13",
"year": "2021"
},
{
"DOI": "10.15252/msb.202110232",
"article-title": "Single-cell analyses reveal SARS-CoV-2 interference with intrinsic immune response in the human gut",
"author": "S Triana",
"doi-asserted-by": "crossref",
"first-page": "e10232",
"issue": "4",
"journal-title": "Mol Syst Biol",
"key": "pbio.3001065.ref030",
"volume": "17",
"year": "2021"
},
{
"article-title": "Long-Term Modeling of SARS-CoV-2 Infection of In Vitro Cultured Polarized Human Airway Epithelium.",
"author": "S Hao",
"issue": "6",
"journal-title": "mBio",
"key": "pbio.3001065.ref031",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1091/mbc.e03-10-0738",
"article-title": "The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response",
"author": "ND Trinklein",
"doi-asserted-by": "crossref",
"first-page": "1254",
"issue": "3",
"journal-title": "Mol Biol Cell",
"key": "pbio.3001065.ref032",
"volume": "15",
"year": "2004"
},
{
"DOI": "10.1126/science.8451637",
"article-title": "Cells in stress: transcriptional activation of heat shock genes",
"author": "RI Morimoto",
"doi-asserted-by": "crossref",
"first-page": "1409",
"issue": "5100",
"journal-title": "Science",
"key": "pbio.3001065.ref033",
"volume": "259",
"year": "1993"
},
{
"DOI": "10.4103/2277-9175.200790",
"article-title": "Identification of Appropriate Housekeeping Genes for Gene Expression Analysis in Long-term Hypoxia-treated Kidney Cells.",
"author": "S Moein",
"doi-asserted-by": "crossref",
"first-page": "15",
"journal-title": "Adv Biomed Res",
"key": "pbio.3001065.ref034",
"volume": "6",
"year": "2017"
},
{
"DOI": "10.1101/gad.293134.116",
"article-title": "High constitutive activity of a broad panel of housekeeping and tissue-specific cis-regulatory elements depends on a subset of ETS proteins",
"author": "A Curina",
"doi-asserted-by": "crossref",
"first-page": "399",
"issue": "4",
"journal-title": "Genes Dev",
"key": "pbio.3001065.ref035",
"volume": "31",
"year": "2017"
},
{
"DOI": "10.1016/j.tig.2013.05.010",
"article-title": "Human housekeeping genes, revisited",
"author": "E Eisenberg",
"doi-asserted-by": "crossref",
"first-page": "569",
"issue": "10",
"journal-title": "Trends Genet",
"key": "pbio.3001065.ref036",
"volume": "29",
"year": "2013"
},
{
"DOI": "10.1074/jbc.AC120.013788",
"article-title": "Inhibition of SARS-CoV-2 by type I and type III interferons",
"author": "U Felgenhauer",
"doi-asserted-by": "crossref",
"first-page": "13958",
"issue": "41",
"journal-title": "J Biol Chem",
"key": "pbio.3001065.ref037",
"volume": "295",
"year": "2020"
},
{
"DOI": "10.1016/j.celrep.2020.107863",
"article-title": "Critical Role of Type III Interferon in Controlling SARS-CoV-2 Infection in Human Intestinal Epithelial Cells",
"author": "ML Stanifer",
"doi-asserted-by": "crossref",
"first-page": "107863",
"issue": "1",
"journal-title": "Cell Rep",
"key": "pbio.3001065.ref038",
"volume": "32",
"year": "2020"
},
{
"DOI": "10.1126/science.abc6027",
"article-title": "Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients",
"author": "J Hadjadj",
"doi-asserted-by": "crossref",
"first-page": "718",
"issue": "6504",
"journal-title": "Science",
"key": "pbio.3001065.ref039",
"volume": "369",
"year": "2020"
},
{
"DOI": "10.1371/journal.pone.0112014",
"article-title": "Inhibitors of the interferon response enhance virus replication in vitro.",
"author": "CE Stewart",
"doi-asserted-by": "crossref",
"first-page": "e112014",
"issue": "11",
"journal-title": "PLoS ONE",
"key": "pbio.3001065.ref040",
"volume": "9",
"year": "2014"
},
{
"DOI": "10.1038/s41375-021-01374-3",
"article-title": "The janus-kinase inhibitor ruxolitinib in SARS-CoV-2 induced acute respiratory distress syndrome (ARDS).",
"author": "A Neubauer",
"doi-asserted-by": "crossref",
"first-page": "2917",
"issue": "10",
"journal-title": "Leukemia",
"key": "pbio.3001065.ref041",
"volume": "35",
"year": "2021"
},
{
"DOI": "10.1128/jvi.2.10.955-961.1968",
"article-title": "Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero).",
"author": "J Desmyter",
"doi-asserted-by": "crossref",
"first-page": "955",
"issue": "10",
"journal-title": "J Virol",
"key": "pbio.3001065.ref042",
"volume": "2",
"year": "1968"
},
{
"article-title": "Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis",
"author": "JD Mosca",
"first-page": "2279",
"issue": "6",
"journal-title": "Mol Cell Biol",
"key": "pbio.3001065.ref043",
"volume": "6",
"year": "1986"
},
{
"DOI": "10.1016/j.cell.2020.04.011",
"article-title": "The Architecture of SARS-CoV-2 Transcriptome.",
"author": "D Kim",
"doi-asserted-by": "crossref",
"first-page": "914",
"issue": "4",
"journal-title": "Cell",
"key": "pbio.3001065.ref044",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1016/j.chom.2020.03.023",
"article-title": "Infection and Rapid Transmission of SARS-CoV-2 in Ferrets.",
"author": "YI Kim",
"doi-asserted-by": "crossref",
"first-page": "704",
"issue": "5",
"journal-title": "Cell Host Microbe",
"key": "pbio.3001065.ref045",
"volume": "27",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.06.011",
"article-title": "A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies",
"author": "AO Hassan",
"doi-asserted-by": "crossref",
"first-page": "744",
"issue": "3",
"journal-title": "Cell",
"key": "pbio.3001065.ref046",
"volume": "182",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2342-5",
"article-title": "Pathogenesis and transmission of SARS-CoV-2 in golden hamsters",
"author": "SF Sia",
"doi-asserted-by": "crossref",
"first-page": "834",
"issue": "7818",
"journal-title": "Nature",
"key": "pbio.3001065.ref047",
"volume": "583",
"year": "2020"
},
{
"DOI": "10.1128/JVI.00957-20",
"article-title": "Replication of Severe Acute Respiratory Syndrome Coronavirus 2 in Human Respiratory Epithelium",
"author": "A Milewska",
"doi-asserted-by": "crossref",
"issue": "15",
"journal-title": "J Virol",
"key": "pbio.3001065.ref048",
"volume": "94",
"year": "2020"
},
{
"DOI": "10.1016/j.stem.2020.10.004",
"article-title": "Three-Dimensional Human Alveolar Stem Cell Culture Models Reveal Infection Response to SARS-CoV-2.",
"author": "J Youk",
"doi-asserted-by": "crossref",
"first-page": "905",
"issue": "6",
"journal-title": "Cell Stem Cell",
"key": "pbio.3001065.ref049",
"volume": "27",
"year": "2020"
},
{
"DOI": "10.1189/jlb.0313153",
"article-title": "Cellular stress response and innate immune signaling: integrating pathways in host defense and inflammation",
"author": "S Muralidharan",
"doi-asserted-by": "crossref",
"first-page": "1167",
"issue": "6",
"journal-title": "J Leukoc Biol",
"key": "pbio.3001065.ref050",
"volume": "94",
"year": "2013"
},
{
"article-title": "Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential.",
"author": "E Kindler",
"first-page": "e00611",
"issue": "1",
"journal-title": "mBioEpub 2013 Feb 21",
"key": "pbio.3001065.ref051",
"volume": "4",
"year": "2013"
},
{
"DOI": "10.1016/j.immuni.2017.03.011",
"article-title": "A Tunable Diffusion-Consumption Mechanism of Cytokine Propagation Enables Plasticity in Cell-to-Cell Communication in the Immune System",
"author": "A Oyler-Yaniv",
"doi-asserted-by": "crossref",
"first-page": "609",
"issue": "4",
"journal-title": "Immunity",
"key": "pbio.3001065.ref052",
"volume": "46",
"year": "2017"
},
{
"DOI": "10.1016/j.molcel.2017.05.011",
"article-title": "Catch and Release of Cytokines Mediated by Tumor Phosphatidylserine Converts Transient Exposure into Long-Lived Inflammation",
"author": "J Oyler-Yaniv",
"doi-asserted-by": "crossref",
"first-page": "635",
"issue": "5",
"journal-title": "Mol Cell",
"key": "pbio.3001065.ref053",
"volume": "66",
"year": "2017"
},
{
"DOI": "10.1371/journal.ppat.1003703",
"article-title": "Defective viral genomes arising in vivo provide critical danger signals for the triggering of lung antiviral immunity",
"author": "K Tapia",
"doi-asserted-by": "crossref",
"first-page": "e1003703",
"issue": "10",
"journal-title": "PLoS Pathog",
"key": "pbio.3001065.ref054",
"volume": "9",
"year": "2013"
},
{
"DOI": "10.1099/jgv.0.000687",
"article-title": "Single-cell studies of IFN-beta promoter activation by wild-type and NS1-defective influenza A viruses",
"author": "MJ Killip",
"doi-asserted-by": "crossref",
"first-page": "357",
"issue": "3",
"journal-title": "J Gen Virol",
"key": "pbio.3001065.ref055",
"volume": "98",
"year": "2017"
},
{
"DOI": "10.1371/journal.ppat.1006650",
"article-title": "Reduced accumulation of defective viral genomes contributes to severe outcome in influenza virus infected patients.",
"author": "J Vasilijevic",
"doi-asserted-by": "crossref",
"first-page": "e1006650",
"issue": "10",
"journal-title": "PLoS Pathog",
"key": "pbio.3001065.ref056",
"volume": "13",
"year": "2017"
},
{
"DOI": "10.1128/JVI.00500-19",
"article-title": "Single-Cell Virus Sequencing of Influenza Infections That Trigger Innate Immunity",
"author": "AB Russell",
"doi-asserted-by": "crossref",
"issue": "14",
"journal-title": "J Virol",
"key": "pbio.3001065.ref057",
"volume": "93",
"year": "2019"
},
{
"DOI": "10.1371/journal.pone.0016546",
"article-title": "Heat shock protein 70 inhibits the activity of Influenza A virus ribonucleoprotein and blocks the replication of virus in vitro and in vivo",
"author": "G Li",
"doi-asserted-by": "crossref",
"first-page": "e16546",
"issue": "2",
"journal-title": "PLoS ONE",
"key": "pbio.3001065.ref058",
"volume": "6",
"year": "2011"
},
{
"DOI": "10.1128/JVI.78.3.1263-1270.2004",
"article-title": "Heat shock protein 70 is related to thermal inhibition of nuclear export of the influenza virus ribonucleoprotein complex",
"author": "E Hirayama",
"doi-asserted-by": "crossref",
"first-page": "1263",
"issue": "3",
"journal-title": "J Virol",
"key": "pbio.3001065.ref059",
"volume": "78",
"year": "2004"
},
{
"DOI": "10.1038/s41392-020-00233-4",
"article-title": "Stress proteins: the biological functions in virus infection, present and challenges for target-based antiviral drug development.",
"author": "Q Wan",
"doi-asserted-by": "crossref",
"first-page": "125",
"issue": "1",
"journal-title": "Signal Transduct Target Ther",
"key": "pbio.3001065.ref060",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.1126/science.aao0409",
"article-title": "An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism",
"author": "P Wang",
"doi-asserted-by": "crossref",
"first-page": "1051",
"issue": "6366",
"journal-title": "Science",
"key": "pbio.3001065.ref061",
"volume": "358",
"year": "2017"
},
{
"DOI": "10.1186/1743-422X-3-58",
"article-title": "Temperature sensitive influenza A virus genome replication results from low thermal stability of polymerase-cRNA complexes",
"author": "RM Dalton",
"doi-asserted-by": "crossref",
"first-page": "58",
"journal-title": "Virol J",
"key": "pbio.3001065.ref062",
"volume": "3",
"year": "2006"
},
{
"DOI": "10.1038/s41579-018-0115-z",
"article-title": "Host and viral determinants of influenza A virus species specificity",
"author": "JS Long",
"doi-asserted-by": "crossref",
"first-page": "67",
"issue": "2",
"journal-title": "Nat Rev Microbiol",
"key": "pbio.3001065.ref063",
"volume": "17",
"year": "2019"
},
{
"DOI": "10.1007/BF01311080",
"article-title": "Characterization of a temperature sensitive feline infectious peritonitis coronavirus",
"author": "KK Christianson",
"doi-asserted-by": "crossref",
"first-page": "185",
"issue": "3–4",
"journal-title": "Arch Virol",
"key": "pbio.3001065.ref064",
"volume": "109",
"year": "1989"
},
{
"DOI": "10.1128/JVI.02140-18",
"article-title": "Analysis of Coronavirus Temperature-Sensitive Mutants Reveals an Interplay between the Macrodomain and Papain-Like Protease Impacting Replication and Pathogenesis",
"author": "X Deng",
"doi-asserted-by": "crossref",
"first-page": "e021140",
"issue": "12",
"journal-title": "J Virol",
"key": "pbio.3001065.ref065",
"volume": "93",
"year": "2019"
},
{
"DOI": "10.1016/B978-012465330-6/50003-8",
"article-title": "2—Virus isolation and quantitation",
"author": "JC Hierholzer",
"doi-asserted-by": "crossref",
"first-page": "25",
"journal-title": "Virology Methods Manual. London: Academic Press",
"key": "pbio.3001065.ref066",
"year": "1996"
},
{
"DOI": "10.1016/S0014-5793(99)01598-7",
"article-title": "The central interactive region of human MxA GTPase is involved in GTPase activation and interaction with viral target structures",
"author": "F Flohr",
"doi-asserted-by": "crossref",
"first-page": "24",
"issue": "1–2",
"journal-title": "FEBS Lett",
"key": "pbio.3001065.ref067",
"volume": "463",
"year": "1999"
},
{
"DOI": "10.1038/nmeth.3317",
"article-title": "HISAT: a fast spliced aligner with low memory requirements.",
"author": "D Kim",
"doi-asserted-by": "crossref",
"first-page": "357",
"issue": "4",
"journal-title": "Nat Methods.",
"key": "pbio.3001065.ref068",
"volume": "12",
"year": "2015"
},
{
"DOI": "10.1093/bioinformatics/btt656",
"article-title": "featureCounts: an efficient general purpose program for assigning sequence reads to genomic features",
"author": "Y Liao",
"doi-asserted-by": "crossref",
"first-page": "923",
"issue": "7",
"journal-title": "Bioinformatics",
"key": "pbio.3001065.ref069",
"volume": "30",
"year": "2014"
},
{
"DOI": "10.1093/bioinformatics/btp616",
"article-title": "edgeR: a Bioconductor package for differential expression analysis of digital gene expression data",
"author": "MD Robinson",
"doi-asserted-by": "crossref",
"first-page": "139",
"issue": "1",
"journal-title": "Bioinformatics",
"key": "pbio.3001065.ref070",
"volume": "26",
"year": "2010"
},
{
"DOI": "10.1093/bioinformatics/btp324",
"article-title": "Fast and accurate short read alignment with Burrows-Wheeler transform",
"author": "H Li",
"doi-asserted-by": "crossref",
"first-page": "1754",
"issue": "14",
"journal-title": "Bioinformatics",
"key": "pbio.3001065.ref071",
"volume": "25",
"year": "2009"
},
{
"DOI": "10.1101/gr.268110.120",
"article-title": "Subgenomic RNA identification in SARS-CoV-2 genomic sequencing data",
"author": "MD Parker",
"doi-asserted-by": "crossref",
"first-page": "645",
"issue": "4",
"journal-title": "Genome Res",
"key": "pbio.3001065.ref072",
"volume": "31",
"year": "2021"
},
{
"DOI": "10.1093/nar/gkx1132",
"article-title": "The Reactome Pathway Knowledgebase",
"author": "A Fabregat",
"doi-asserted-by": "crossref",
"first-page": "D649",
"issue": "D1",
"journal-title": "Nucleic Acids Res",
"key": "pbio.3001065.ref073",
"volume": "46",
"year": "2018"
},
{
"article-title": "The reactome pathway knowledgebase",
"author": "B Jassal",
"first-page": "D498",
"issue": "D1",
"journal-title": "Nucleic Acids Res",
"key": "pbio.3001065.ref074",
"volume": "48",
"year": "2020"
},
{
"DOI": "10.1038/s41467-019-09234-6",
"article-title": "Metascape provides a biologist-oriented resource for the analysis of systems-level datasets.",
"author": "Y Zhou",
"doi-asserted-by": "crossref",
"first-page": "1523",
"issue": "1",
"journal-title": "Nat Commun",
"key": "pbio.3001065.ref075",
"volume": "10",
"year": "2019"
},
{
"DOI": "10.1186/s13059-018-1618-7",
"article-title": "An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar",
"author": "ND Grubaugh",
"doi-asserted-by": "crossref",
"first-page": "8",
"issue": "1",
"journal-title": "Genome Biol",
"key": "pbio.3001065.ref076",
"volume": "20",
"year": "2019"
}
],
"reference-count": 76,
"references-count": 76,
"relation": {},
"resource": {
"primary": {
"URL": "https://dx.plos.org/10.1371/journal.pbio.3001065"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"General Agricultural and Biological Sciences",
"General Immunology and Microbiology",
"General Biochemistry, Genetics and Molecular Biology",
"General Neuroscience"
],
"subtitle": [],
"title": "Elevated temperature inhibits SARS-CoV-2 replication in respiratory epithelium independently of IFN-mediated innate immune defenses",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1371/journal.pbio.corrections_policy",
"update-to": [
{
"DOI": "10.1371/journal.pbio.3001065",
"label": "New version",
"type": "new_version",
"updated": {
"date-parts": [
[
2022,
1,
18
]
],
"date-time": "2022-01-18T00:00:00Z",
"timestamp": 1642464000000
}
}
],
"volume": "19"
}