Identifying DNA Methylation Patterns in Post COVID-19 Condition: Insights from a One-Year Prospective Cohort Study
et al., medRxiv, doi:10.1101/2025.02.28.25323075, CoVUm, NCT04368013, Mar 2025
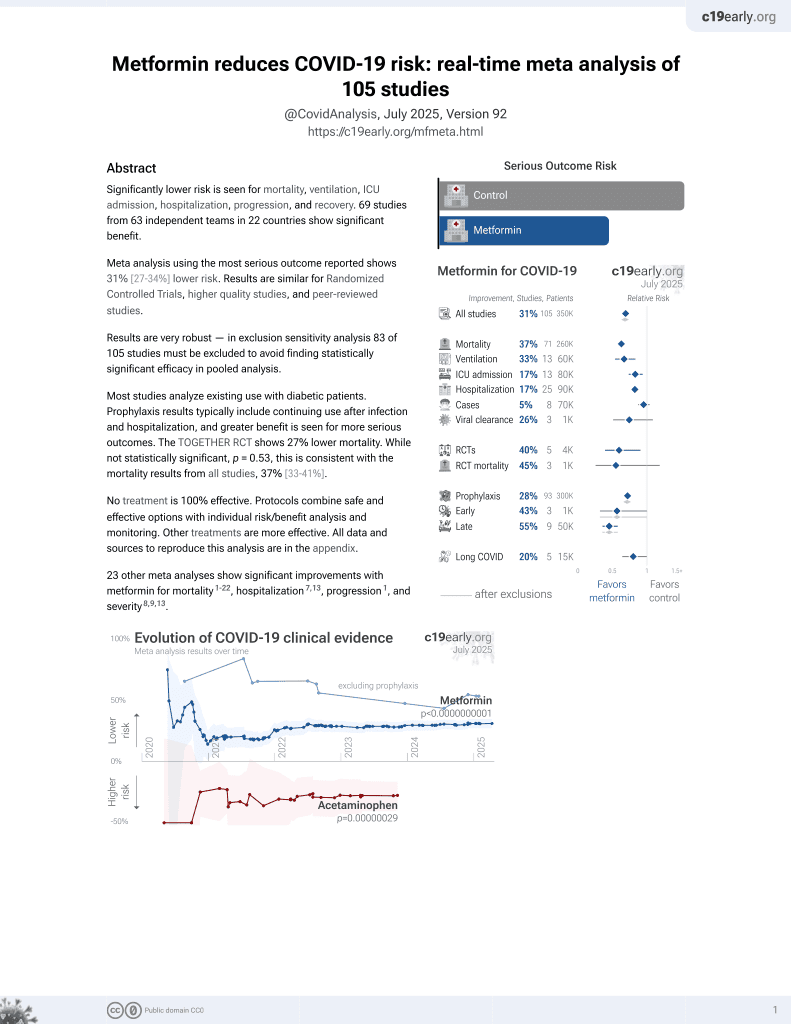
Metformin for COVID-19
3rd treatment shown to reduce risk in
July 2020, now with p < 0.00000000001 from 110 studies.
Lower risk for mortality, ventilation, ICU, hospitalization, progression, recovery, and viral clearance.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
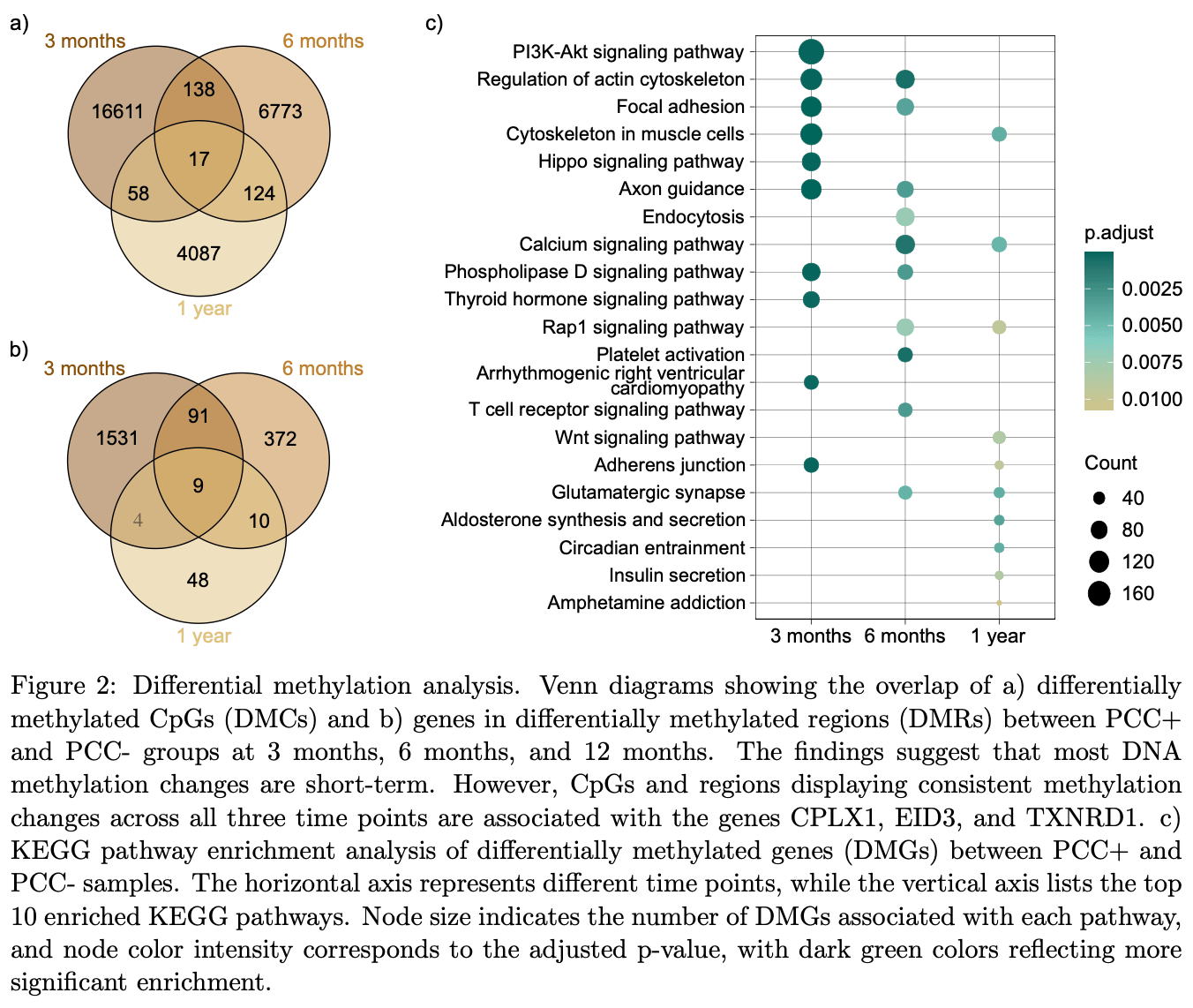
Prospective cohort study with 22 Post COVID-19 condition (PCC+) patients and 22 matched COVID-19 convalescents (PCC-), showing distinct DNA methylation patterns diminishing over time. The study identified TXNRD1 methylation changes associated with cognitive symptoms and fatigue, implicating redox imbalance in PCC pathology. Pathway analysis revealed enrichment in PI3K-Akt and AMPK signaling pathways, potentially explaining the observed efficacy of metformin in reducing PCC incidence.
Granvik et al., 4 Mar 2025, prospective, Sweden, preprint, 12 authors, trial NCT04368013 (history) (CoVUm).
Contact: christoffer.granvik@umu.se, jelena.smiljanic@umu.se, alicia.lind@umu.se, david.martinez@liu.se, shumaila.sayyab@liu.se, clas.ahlm@umu.se, mattias.forsell@umu.se, sara.cajander@oru.se, mika.gustafsson@liu.se, martin.rosvall@umu.se, johan.normark@umu.se, maria.lerm@liu.se.
Identifying DNA Methylation Patterns in Post COVID-19 Condition: Insights from a One-Year Prospective Cohort Study
doi:10.1101/2025.02.28.25323075
The mechanisms underlying Post COVID-19 condition (PCC), with its range of long-lasting symptoms, remain unclear. This study investigates DNA methylation patterns over one year in a subset of nonhospitalized COVID-19 patients with persistent symptoms and reduced quality of life, termed PCC+ (Post COVID-19 condition plus). In a cohort of 22 PCC+ individuals and 22 matched COVID-19 convalescents (PCC-), we identified distinct DNA methylation differences between the groups that diminish over time. Methylation changes in the TXNRD1 gene were significantly associated with cognitive symptoms and fatigue, implicating redox imbalance in PCC pathology. Pathway analysis revealed enrichment in PI3K-Akt and AMPK signaling pathways, potentially underlying the observed efficacy of metformin in reducing PCC incidence. While we found no differences in epigenetic age acceleration between the groups, we observed longitudinal changes in the methylation of the RAS and RAP1 signaling pathways. These findings provide crucial insights into PCC+ mechanisms and suggest oxidative stress pathways as promising targets for therapeutic interventions.
Ethical considerations The study was approved by the Swedish Ethical Review Authority (approval number: 2020-01557) and was carried out according to the declaration of Helsinki.
Data Availability Statement The datasets presented in this article are not readily available because ethical approval does not support publication of the entire dataset of the study cohort. Requests to access the datasets should be directed to johan.normark@umu.se.
Funding The study was supported by grants from SciLifeLab, National COVID-19 Research Program (VC-2020-0015), financed by the Knut and Alice Wallenberg Foundation, Swedish Heart-Lung Foundation (20200325, 202100789 and 20220325).
References
Ahmad, Edin, Granvik, Kumm Persson, Tevell et al., High prevalence of persistent symptoms and reduced health-related quality of life 6 months after covid-19, Frontiers in Public Health
André, Antunes, Reis-De Oliveira, Martins, Souza, Molecular overlaps of neurological manifestations of covid-19 and schizophrenia from a proteomic perspective, European Archives of Psychiatry and Clinical Neuroscience
Anil, Ori, Ake, Lu, Horvath et al., Significant variation in the performance of dna methylation predictors across data preprocessing and normalization strategies, Genome Biology
Aryee, Jaffe, Corrada-Bravo, Ladd-Acosta, Feinberg et al., Minfi: a flexible and comprehensive bioconductor package for the analysis of infinium dna methylation microarrays, Bioinformatics
Bahmer, Borzikowsky, Lieb, Horn, Krist et al., Severity, predictors and clinical correlates of post-covid syndrome (pcs) in germany: A prospective, multi-centre, population-based cohort study, eClinicalMedicine
Balnis, Madrid, Drake, Vancavage, Tiwari et al., Blood dna methylation in post-acute sequelae of covid-19 (pasc): a prospective cohort study
Bramante, Buse, Liebovitz, Nicklas, Puskarich et al., Outpatient treatment of covid-19 and incidence of post-covid-19 condition over 10 months (covid-out): a multicentre, randomised, quadruple-blind, parallel-group, phase 3 trial, The Lancet Infectious Diseases
Calzari, Fernando, Dragani, Zanotti, Inglese et al., Epigenetic patterns, accelerated biological aging, and enhanced epigenetic drift detected 6 months following covid-19 infection: insights from a genome-wide dna methylation study, Clinical Epigenetics
Cao, Li, Wang, Ran, Davalos et al., Accelerated biological aging in covid-19 patients, Nature Communications
Cervia-Hasler, Brüningk, Hoch, Fan, Muzio et al., Persistent complement dysregulation with signs of thromboinflammation in active long covid, Science
Cherra, Lamb, Interactions between ras and rap signaling pathways during neurodevelopment in health and disease, Frontiers in molecular neuroscience
David R Anderson, Burnham, Avoiding pitfalls when using information-theoretic methods, The Journal of wildlife management
Fan, Yang, Tian, The nmda receptor complex: a multifunctional machine at the glutamatergic synapse, Frontiers in cellular neuroscience
Gallardo, Todd, Lima, Chekan, Chiu et al., Sars-cov-2 main protease targets host selenoproteins and glutathione biosynthesis for knockdown via proteolysis, potentially disrupting the thioredoxin and glutaredoxin redox cycles, Antioxidants
Gay, Desquiret-Dumas, Nagot, Rapenne, Van De Perre et al., Long-term persistence of mitochondrial dysfunctions after viral infections and antiviral therapies: A review of mechanisms involved, Journal of medical virology
Golin, Tinkov, Aschner, Farina, Batista Teixeira Da Rocha, Relationship between selenium status, selenoproteins and covid-19 and other inflammatory diseases: A critical review, Journal of Trace Elements in Medicine and Biology
Hannah E Davis, Mccorkell, Vogel, Topol, Long covid: major findings, mechanisms and recommendations, Nature Reviews Microbiology
Hannum, Guinney, Zhao, Zhang, Hughes et al., Genome-wide Methylation Profiles Reveal Quantitative Views of Human Aging Rates, Molecular Cell
Hao, Zhao, Towers, Liao, Monteiro et al., Txnrd1 drives the innate immune response in senescent cells with implications for age-associated inflammation, Nature Aging
Harris, Taylor, Thielke, Payne, Gonzalez et al., Research electronic data capture (redcap)-a metadata-driven methodology and workflow process for providing translational research informatics support, Journal of Biomedical Informatics
Ho, Johnson, Tarapore, Janakiram, Zhang et al., Environmental epigenetics and its implication on disease risk and health outcomes, ILAR journal
Horvath, DNA methylation age of human tissues and cell types, Genome Biology
Horvath, Oshima, Martin, Lu, Quach et al., Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies, Aging
Hou, Song, Li, Zhang, Wang et al., Metformin reduces intracellular reactive oxygen species levels by upregulating expression of the antioxidant thioredoxin via the ampk-foxo3 pathway, Biochemical and Biophysical Research Communications
Huoman, Sayyab, Apostolou, Karlsson, Porcile et al., Epigenetic rewiring of pathways related to odour perception in immune cells exposed to sars-cov-2 in vivo and in vitro, Epigenetics
Iain R Konigsberg, Barnes, Campbell, Davidson, Zhen et al., Host methylation predicts sars-cov-2 infection and clinical outcome, Communications medicine
John, Marshall, A minimal common outcome measure set for covid-19 clinical research, The Lancet Infectious Diseases
Johnson, Li, Rabinovic, Adjusting batch effects in microarray expression data using empirical bayes methods, Biostatistics
Kadhem Al-Hakeim, Tahseen Al-Rubaye, Shihab Al-Hadrawi, Almulla, Maes, Long-covid post-viral chronic fatigue and affective symptoms are associated with oxidative damage, lowered antioxidant defenses and inflammation: a proof of concept and mechanism study, Molecular Psychiatry
Khallaghi, Safarian, Nasoohi, Ahmadiani, Dargahi, Metformin-induced protection against oxidative stress is associated with akt/mtor restoration in pc12 cells, Life Sciences
Kosuru, Singh, Lakshmikanthan, Nishijima, Vasquez-Vivar et al., Distinct signaling functions of rap1 isoforms in no release from endothelium
Lasalle, Dna methylation biomarkers of intellectual/developmental disability across the lifespan, Journal of Neurodevelopmental Disorders
Lee, Riskedal, Kalleberg, Istre, Lind et al., Ewas of post-covid-19 patients shows methylation differences in the immune-response associated gene, ifi44l, three months after covid-19 infection, Scientific Reports
Levine, Lu, Quach, Chen, Themistocles et al., An epigenetic biomarker of aging for lifespan and healthspan, Aging
Martínez-Enguita, Sanjiv, Dwivedi, Jörnsten, Gustafsson, NCAE: data-driven representations using a deep network-coherent DNA methylation autoencoder identify robust disease and risk factor signatures, Briefings in Bioinformatics
Nikesjö, Sayyab, Eirini Apostolou, Rosén, Hedman et al., Defining post-acute covid-19 syndrome (pacs) by an epigenetic biosignature in peripheral blood mononuclear cells, Clinical Epigenetics
Nishiyama, Taniguchi, Kondoh, Kimura, Kato et al., A simple assessment of dyspnoea as a prognostic indicator in idiopathic pulmonary fibrosis, European Respiratory Journal
O'mahoney, Routen, Gillies, Ekezie, Welford et al., The prevalence and long-term health effects of long covid among hospitalised and non-hospitalised populations: a systematic review and meta-analysis, eClinicalMedicine
Peters, Buckley, Statham, Pidsley, Samaras et al., De novo identification of differentially methylated regions in the human genome, Epigenetics & chromatin
Phetsouphanh, David R Darley, Daniel B Wilson, Howe, Munier et al., Immunological dysfunction persists for 8 months following initial mild-to-moderate sars-cov-2 infection, Nature immunology
Phipson, Maksimovic, Oshlack, missmethyl: an r package for analyzing data from illumina's humanmethylation450 platform, Bioinformatics
Ritchie, Phipson, Wu, Hu, Law et al., limma powers differential expression analyses for rna-sequencing and microarray studies, Nucleic Acids Research
Roffman, Allison, Charlson comorbidities index, Journal of physiotherapy
Schomburg, Selenium deficiency in covid-19-a possible long-lasting toxic relationship, Nutrients
Shan V Andrews, Sheppard, Windham, Schieve, Schendel et al., Case-control meta-analysis of blood dna methylation and autism spectrum disorder, Molecular autism
Shijie C Zheng, Breeze, Beck, Teschendorff, Identification of differentially methylated cell types in epigenome-wide association studies, Nature methods
Who, A clinical case definition of post covid-19 condition by a delphi consensus
Wu, Hu, Xu, Chen, Guo et al., clusterprofiler 4.0: A universal enrichment tool for interpreting omics data, The innovation
Xu, Niu, Li, Taylor, Enmix: a novel background correction method for illumina humanmethylation450 beadchip, Nucleic Acids Research
Yuan Tian, Morris, Webster, Yang, Beck et al., Champ: updated methylation analysis pipeline for illumina beadchips, Bioinformatics
Zhang, Li, Ding, Guo, Ma, Rap1b: A cytoskeletal regulator advantageous to viral infection, iScience
Zhang, Vallerga, Walker, Lin, Henders et al., Improved precision of epigenetic clock estimates across tissues and its implication for biological ageing, Genome Medicine
DOI record:
{
"DOI": "10.1101/2025.02.28.25323075",
"URL": "http://dx.doi.org/10.1101/2025.02.28.25323075",
"abstract": "<jats:p>The mechanisms underlying Post COVID-19 condition (PCC), with its range of long-lasting symptoms, remain unclear. This study investigates DNA methylation patterns over one year in a subset of non-hospitalized COVID-19 patients with persistent symptoms and reduced quality of life, termed PCC+ (Post COVID-19 condition plus). In a cohort of 22 PCC+ individuals and 22 matched COVID-19 convalescents (PCC-), we identified distinct DNA methylation differences between the groups that diminish over time. Methylation changes in the TXNRD1 gene were significantly associated with cognitive symptoms and fatigue, implicating redox imbalance in PCC pathology. Pathway analysis revealed enrichment in PI3K-Akt and AMPK signaling pathways, potentially underlying the observed efficacy of metformin in reducing PCC incidence. While we found no differences in epigenetic age acceleration between the groups, we observed longitudinal changes in the methylation of the RAS and RAP1 signaling pathways. These findings provide crucial insights into PCC+ mechanisms and suggest oxidative stress pathways as promising targets for therapeutic interventions.</jats:p>",
"accepted": {
"date-parts": [
[
2025,
3,
4
]
]
},
"author": [
{
"ORCID": "https://orcid.org/0009-0001-5257-8109",
"affiliation": [],
"authenticated-orcid": false,
"family": "Granvik",
"given": "Christoffer",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0000-0003-0124-1909",
"affiliation": [],
"authenticated-orcid": false,
"family": "Smiljanic",
"given": "Jelena",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-0309-1852",
"affiliation": [],
"authenticated-orcid": false,
"family": "Lind",
"given": "Alicia",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-6363-6298",
"affiliation": [],
"authenticated-orcid": false,
"family": "Martinez-Enguita",
"given": "David",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-4127-4887",
"affiliation": [],
"authenticated-orcid": false,
"family": "Sayyab",
"given": "shumaila",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-2018-8592",
"affiliation": [],
"authenticated-orcid": false,
"family": "Ahlm",
"given": "Clas",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-6904-742X",
"affiliation": [],
"authenticated-orcid": false,
"family": "Forsell",
"given": "Mattias",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-3921-4244",
"affiliation": [],
"authenticated-orcid": false,
"family": "Cajander",
"given": "Sara",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-0048-4063",
"affiliation": [],
"authenticated-orcid": false,
"family": "Gustafsson",
"given": "Mika",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-7181-9940",
"affiliation": [],
"authenticated-orcid": false,
"family": "Rosvall",
"given": "Martin",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-5831-4369",
"affiliation": [],
"authenticated-orcid": false,
"family": "Normark",
"given": "Johan",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-5092-9892",
"affiliation": [],
"authenticated-orcid": false,
"family": "Lerm",
"given": "Maria",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
3,
4
]
],
"date-time": "2025-03-04T18:00:13Z",
"timestamp": 1741111213000
},
"deposited": {
"date-parts": [
[
2025,
3,
4
]
],
"date-time": "2025-03-04T18:00:13Z",
"timestamp": 1741111213000
},
"group-title": "Infectious Diseases (except HIV/AIDS)",
"indexed": {
"date-parts": [
[
2025,
3,
4
]
],
"date-time": "2025-03-04T18:40:29Z",
"timestamp": 1741113629192,
"version": "3.38.0"
},
"institution": [
{
"name": "medRxiv"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2025,
3,
4
]
]
},
"license": [
{
"URL": "http://creativecommons.org/licenses/by-nc/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
3,
4
]
],
"date-time": "2025-03-04T00:00:00Z",
"timestamp": 1741046400000
}
}
],
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2025.02.28.25323075",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2025,
3,
4
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2025,
3,
4
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "http://medrxiv.org/lookup/doi/10.1101/2025.02.28.25323075"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"subtype": "preprint",
"title": "Identifying DNA Methylation Patterns in Post COVID-19 Condition: Insights from a One-Year Prospective Cohort Study",
"type": "posted-content"
}