Single cell sequencing reveals cellular landscape alterations in the airway mucosa of patients with pulmonary long COVID
et al., medRxiv, doi:10.1101/2024.02.26.24302674, Feb 2024
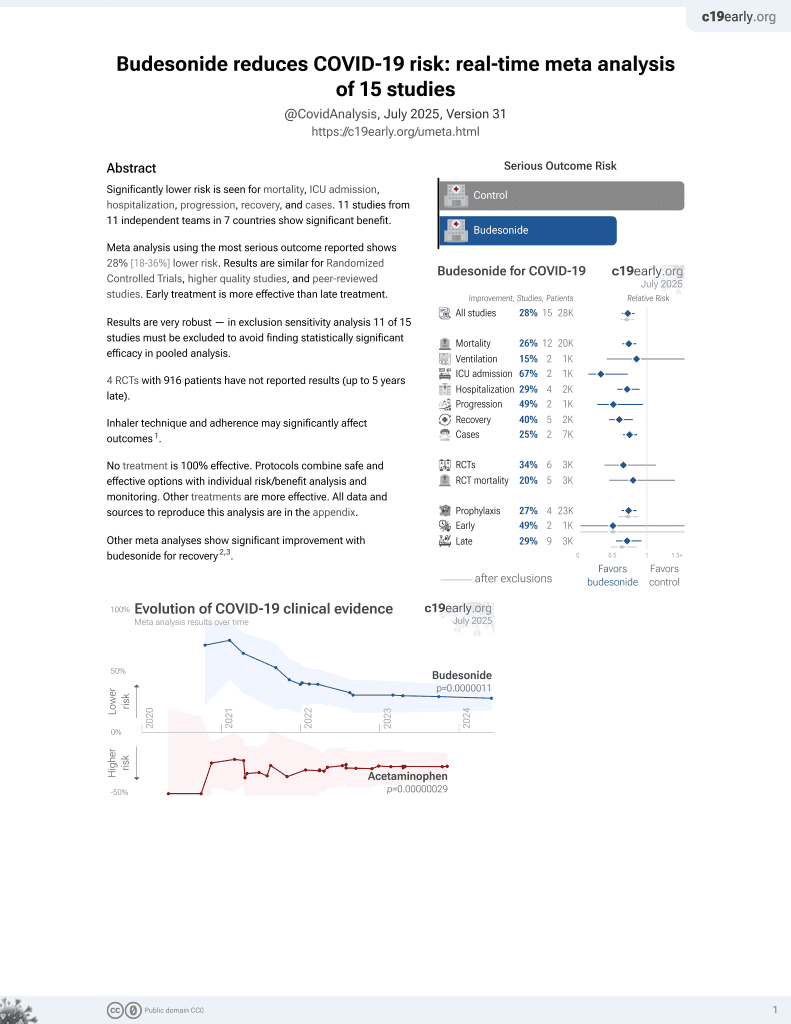
Budesonide for COVID-19
28th treatment shown to reduce risk in
September 2021, now with p = 0.0000042 from 14 studies, recognized in 10 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
Single-cell RNA sequencing study of airway cells from patients with persistent respiratory symptoms due to COVID-19 infection (pulmonary long COVID) over 1 year after initial infection. Authors found increased neutrophils and neutrophil activation signatures in pulmonary long COVID airway cells compared to controls. Ingenuity pathway analysis predicted that budesonide could potentially revert the airway cell gene expression signature of pulmonary long COVID patients to resemble that of controls.
Gerayeli et al., 28 Feb 2024, preprint, 18 authors.
Contact: don.sin@hli.ubc.ca.
Ex vivo studies are an important part of preclinical research, however results may be very different in vivo.
Single cell sequencing reveals cellular landscape alterations in the airway mucosa of patients with pulmonary long COVID
doi:10.1101/2024.02.26.24302674
Single cell profiling shows the infiltration of neutrophils with upregulation of mucin genes in the airway mucosa of patients with pulmonary long COVID, indicating persistent small airway inflammation in pulmonary long COVID.
Author Contributions: FVG, XL,CXY ran the analysis and co-wrote the method section. JT and GK analyzed the IPA analysis and contributed to the method section & discussion section. FVG, HYP, SM,XL,CXY,CYC,JT ,GK and DDS were involved in interpretation of the results. FVG,SM,CYC,EG,RLD,CG,JSWY,TS, DA,TS,JML contributed to the experiment and data acquisition. FVG, HYP and DDS prepared the first draft of the manuscript. All authors contributed to revise the manuscript and approved the final draft of the manuscript.
Supplemental Figures: Supplemental Figure S1A-C
References
Chen, Sang, Jiang, Longitudinal hematologic and immunologic variations associated with the progression of COVID-19 patients in China, J Allergy Clin Immunol
Chiang, Korinek, Cheng, Targeting Neutrophils to Treat Acute Respiratory Distress Syndrome in Coronavirus Disease, Front Pharmacol
Davis, Assaf, Mccorkell, Characterizing long COVID in an international cohort: 7 months of symptoms and their impact, EClinicalMedicine
Davis, Mccorkell, Vogel, Long COVID: major findings, mechanisms and recommendations, Nat Rev Microbiol
Franzen, Gan, Bjorkegren, PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data, Database
Gentilotti, Gorska, Tami, Clinical phenotypes and quality of life to define post-COVID-19 syndrome: a cluster analysis of the multinational, prospective ORCHESTRA cohort, EClinicalMedicine
George, Reed, Desai, A persistent neutrophil-associated immune signature characterizes post-COVID-19 pulmonary sequelae, Sci Transl Med
Gerayeli, Milne, Yang, Single-cell RNA sequencing of bronchoscopy specimens: development of a rapid minimal handling protocol, Biotechniques
Greenhalgh, Knight, Court, Management of post-acute covid-19 in primary care, BMJ
Hewitt, Lloyd, Regulation of immune responses by the airway epithelial cell landscape, Nat Rev Immunol
Li, Zhang, Zhang, Neutrophils in COVID-19: recent insights and advances, Virol J
Mehandru, Merad, Pathological sequelae of long-haul COVID, Nat Immunol
Milne, Li, Yang, Inhaled corticosteroids downregulate SARS-CoV-2related genes in COPD: results from a randomised controlled trial, Eur Respir J
Proal, Vanelzakker, Long COVID or Post-acute Sequelae of COVID-19 (PASC): An Overview of Biological Factors That May Contribute to Persistent Symptoms, Front Microbiol
Schulte-Schrepping, Reusch, Paclik, Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment, Cell
Sekine, Perez-Potti, Rivera-Ballesteros, Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19, Cell
Sun, Sun, Prolonged Persistence of SARS-CoV-2 RNA in Body Fluids, Emerg Infect Dis
Tanabe, Fujimoto, Yasuo, Modulation of mucus production by interleukin-13 receptor alpha2 in the human airway epithelium, Clin Exp Allergy
Wilk, Rustagi, Zhao, A single-cell atlas of the peripheral immune response in patients with severe COVID-19, Nat Med
Woodruff, Bonham, Anam, Chronic inflammation, neutrophil activity, and autoreactivity splits long COVID, Nat Commun
Young, Behjati, SoupX removes ambient RNA contamination from dropletbased single-cell RNA sequencing data, Gigascience
Zaragosi, Deprez, Barbry, Using single-cell RNA sequencing to unravel cell lineage relationships in the respiratory tract, Biochem Soc Trans
Zhou, Ren, Zhang, Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients, Cell Host Microbe
DOI record:
{
"DOI": "10.1101/2024.02.26.24302674",
"URL": "http://dx.doi.org/10.1101/2024.02.26.24302674",
"abstract": "<jats:p>To elucidate the important cellular and molecular drivers of pulmonary long COVID, we generated a single-cell transcriptomic map of the airway mucosa using bronchial brushings from patients with long COVID who reported persistent pulmonary symptoms. Adults with and without long COVID were recruited from the general community in greater Vancouver, Canada. The cohort was divided into those with pulmonary long COVID (PLC), which was defined as persons with new or worsening respiratory symptoms following at least one year from their initial acute SARS-CoV-2 infection (N=9); and control subjects defined as SARS-CoV-2 infected persons whose acute respiratory symptoms had fully resolved or individuals who had not experienced acute COVID-19 (N=9). These participants underwent bronchoscopy from which a single cell suspension was created from bronchial brush samples and then sequenced. A total of 56,906 cells were recovered for the downstream analysis, with 34,840 cells belonging to the PLC group. A dimensionality reduction plot shows a unique cluster of neutrophils in the PLC group (p<.05). Ingenuity Pathway Analysis revealed that neutrophil degranulation pathway was enriched across epithelial cells. Differential gene expression analysis between the PLC and control groups demonstrated upregulation of mucin genes in secretory cell clusters. A single-cell transcriptomic landscape of the small airways shows that the PLC airways harbors a dominant neutrophil cluster and an upregulation in the neutrophil-associated activation signature with increased expression of MUC genes in the secretory cells. Together, they suggest that pulmonary symptoms of long COVID may be driven by chronic small airway inflammation.</jats:p>",
"accepted": {
"date-parts": [
[
2024,
2,
28
]
]
},
"author": [
{
"affiliation": [],
"family": "V.Gerayeli",
"given": "Firoozeh",
"sequence": "first"
},
{
"affiliation": [],
"family": "Park",
"given": "Hye Yun",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Milne",
"given": "Stephen",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Li",
"given": "Xuan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Yang",
"given": "Chen Xi",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Tuong",
"given": "Josie",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Eddy",
"given": "Rachel",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Guinto",
"given": "Elizabeth",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Cheung",
"given": "Chung Y",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Yang",
"given": "Julia Shun-Wei",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Gilchrist",
"given": "Cassie",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Yehia",
"given": "Dina",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Stach",
"given": "Tara",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Shaipanich",
"given": "Tawimas",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Leipsic",
"given": "Jonathon",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Koelwyn",
"given": "Graeme",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Leung",
"given": "Janice",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Sin",
"given": "Don D",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
2,
28
]
],
"date-time": "2024-02-28T18:55:31Z",
"timestamp": 1709146531000
},
"deposited": {
"date-parts": [
[
2024,
2,
28
]
],
"date-time": "2024-02-28T18:55:34Z",
"timestamp": 1709146534000
},
"group-title": "Respiratory Medicine",
"indexed": {
"date-parts": [
[
2024,
2,
29
]
],
"date-time": "2024-02-29T00:33:58Z",
"timestamp": 1709166838799
},
"institution": [
{
"name": "medRxiv"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2024,
2,
28
]
]
},
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2024.02.26.24302674",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2024,
2,
28
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2024,
2,
28
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "http://medrxiv.org/lookup/doi/10.1101/2024.02.26.24302674"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subtitle": [],
"subtype": "preprint",
"title": "Single cell sequencing reveals cellular landscape alterations in the airway mucosa of patients with pulmonary long COVID",
"type": "posted-content"
}