Detecting the most critical clinical variables of COVID-19 breakthrough infection in vaccinated persons using machine learning
et al., DIGITAL HEALTH, doi:10.1177/20552076231207593, Nov 2023
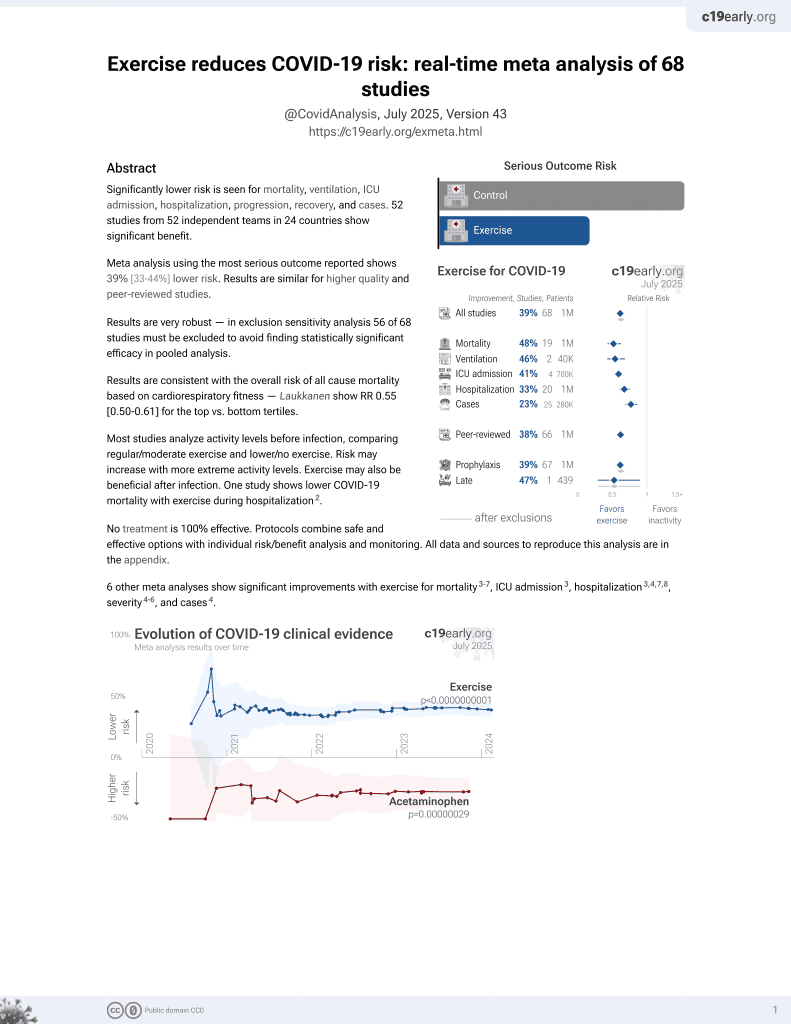
Exercise for COVID-19
9th treatment shown to reduce risk in
October 2020, now with p < 0.00000000001 from 68 studies.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
Retrospective 257 individuals, finding higher physical activity as one of the most predictive variables for lower risk of COVID-19 breakthrough infection.
Daramola et al., 5 Nov 2023, South Africa, peer-reviewed, 14 authors, study period 14 April, 2021 - 3 July, 2021.
Contact: daramolaj@cput.ac.za.
Detecting the most critical clinical variables of COVID-19 breakthrough infection in vaccinated persons using machine learning
DIGITAL HEALTH, doi:10.1177/20552076231207593
Background: COVID-19 vaccines offer different levels of immune protection but do not provide 100% protection. Vaccinated persons with pre-existing comorbidities may be at an increased risk of SARS-CoV-2 breakthrough infection or reinfection. The aim of this study is to identify the critical variables associated with a higher probability of SARS-CoV-2 breakthrough infection using machine learning. Methods: A dataset comprising symptoms and feedback from 257 persons, of whom 203 were vaccinated and 54 unvaccinated, was used for the investigation. Three machine learning algorithms -Deep Multilayer Perceptron (Deep MLP), XGBoost, and Logistic Regressionwere trained with the original (imbalanced) dataset and the balanced dataset created by using the Random Oversampling Technique (ROT), and the Synthetic Minority Oversampling Technique (SMOTE). We compared the performance of the classification algorithms when the features highly correlated with breakthrough infection were used and when all features in the dataset were used.
Result: The results show that when highly correlated features were considered as predictors, with Random Oversampling to address data imbalance, the XGBoost classifier has the best performance (F1 = 0.96; accuracy = 0.96; AUC = 0.98; G-Mean = 0.98; MCC = 0.88). The Deep MLP had the second best performance (F1 = 0.94; accuracy = 0.94; AUC = 0.92; G-Mean = 0.70; MCC = 0.42), while Logistic Regression had less accurate performance (F1 = 0.89; accuracy = 0.88; AUC = 0.89; G-Mean = 0.89; MCC = 0.68). We also used Shapley Additive Explanations (SHAP) to investigate the interpretability of the models. We found that body temperature, total cholesterol, glucose level, blood pressure, waist circumference, body weight, body mass index (BMI), haemoglobin level, and physical activity per week are the most critical variables indicating a higher risk of breakthrough infection.
Conclusion: These results, evident from our unique data source derived from apparently healthy volunteers with cardiovascular risk factors, follow the expected pattern of positive or negative correlations previously reported in the literature. This information strengthens the body of knowledge currently applied in public health guidelines and may also be used by medical practitioners in the future to reduce the risk of SARS-CoV-2 breakthrough infection.
complications from SARS-CoV-2. Woods et al. 25 opined that the immobilisation and the physical inactivity of patients could down-regulate the ability of organ systems to resist viral infection and increase the risk of damage to the immune, respiratory, cardiovascular, and musculoskeletal systems and the brain. Shahidi et al. 42 concurred with Woods et al. 25 by reporting that physical activity benefits include musculoskeletal and cardiovascular health, healthy
References
Abu-Farha, Al-Mulla, Thanaraj, Impact of diabetes in patients diagnosed with COVID-19, Front Immunol
Afrash, Kazemi-Arpanahi, Shanbehzadeh, Predicting hospital readmission risk in patients with COVID-19: a machine learning approach, Informatics Med Unlocked
Ahamad, Aktar, Uddin, Adverse effects of COVID-19 vaccination: machine learning and statistical approach to identify and classify incidences of morbidity and postvaccination reactogenicity, Healthcare
Aparisi, Iglesias-Echeverría, Ybarra-Falcón, Low-density lipoprotein cholesterol levels are associated with poor clinical outcomes in COVID-19, Nutr Metab Cardiovasc Dis
Basso, Negro, Cegolon, Risk of vaccine breakthrough SARS-CoV-2 infection and associated factors in healthcare workers of Trieste teaching hospitals (north-eastern Italy), Viruses
Bornstein, Rubino, Khunti, Practical recommendations for the management of diabetes in patients with COVID-19, Lancet Diabetes Endocrinol
Chawla, Bowyer, Hall, SMOTE: synthetic minority over-sampling technique, J Artif Intell Res
Chen, Guestrin, Xgboost: a scalable tree boosting system
Chen, Ouyang, Bao, An interpretable machine learning framework for accurate severe vs non-severe COVID-19 clinical type classification, SSRN Electron J, doi:10.2139/ssrn.3638427
Chen, Zeng, Yang, Nomogram model for prediction of SARS-CoV-2 breakthrough infection in fujian: a casecontrol real-world study, Front Cell Infect Microbiol
Chicco, Jurman, The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation, BMC Genomics
Daramola, Nyasulu, Mashamba-Thompson, Towards AI-enabled multimodal diagnostics and management of COVID-19 and comorbidities in resource-limited settings, Informatics (MDPI)
Davies, Morden, Rousseau, Outcomes of laboratory-confirmed SARS-CoV-2 infection during resurgence driven by Omicron lineages BA. 4 and BA. 5 compared with previous waves in the Western Cape Province, South Africa, Int J Infect Dis
Dreiseitl, Logistic regression and artificial neural network classification models: a methodology review, J Biomed Inform
Ebrahimi, Sharifi, Mousavi-Roknabadi, Predictive determinants of overall survival among re-infected COVID-19 patients using the elastic-net regularised Cox proportional hazards model: a machine-learning algorithm, BMC Public Health
Eom, Byeon, Predicting the sentiment of South Korean Twitter users toward vaccination after the emergence of COVID-19 Omicron variant using deep learningbased natural language processing, Front Med
Fernández, García, Galar, Foundations on imbalanced classification, doi:10.1007/978-3-319-98074-4_2
Goldreich, Computational complexity: a conceptual perspective, ACM Sigact News
Kočar, Režen, Rozman, Cholesterol, lipoproteins, and COVID-19: basic concepts and clinical applications, Biochim Biophys Acta Mol Cell Biol Lipids
Kotze, Van Rensburg, Davids, Translating population risk into personal utility using a mobile phone app for application of genomic medicine integrating service and research in the COVID-19 era, Clin Chem Lab Med
Kubat, Addressing the curse of imbalanced training sets: one-sided selection, Proc 14th Int Conf Machine Learn
Kulkarni, Pino, French, Real-time anomaly detection framework for many-core router through machinelearning techniques, ACM J Emerging Technol Comput Syst (JETC)
Kumar, Gupta, Agarwal, Epidemic efficacy of Covid-19 vaccination against Omicron: an innovative approach using enhanced residual recurrent neural network, Plos One
Kurano, Ohmiya, Kishi, Measurement of SARS-CoV-2 antibody titers improves the prediction accuracy of COVID-19 maximum severity by machine learning in non-vaccinated patients, Front Immunol
Liao, Song, Ren, VOC-DL: deep learning prediction model for COVID-19 based on VOC virus variants, Comput Methods Programs Biomed
Lundberg, Lee, A unified approach to interpreting model predictions
Muhammad, Algehyne, Usman, Supervised machine learning models for prediction of COVID-19 infection using epidemiology dataset, SN Computer Science
Nusinovici, Tham, Yan, Logistic regression was as good as machine learning for predicting major chronic diseases, J Clin Epidemiol
Olson, Cava, Mustahsan, Data-driven advice for applying machine learning to bioinformatics problems
Park, Lee, Machine learning-based COVID-19 patients triage algorithm using patient-generated health data from nationwide multicenter database, Infect Dis Ther
Radenkovic, Chawla, Pirro, Cholesterol in relation to COVID-19: should we care about it?, J Clin Med
Rashed, Kodera, Hirata, COVID-19 forecasting using new viral variants and vaccination effectiveness models, Comput Biol Med
Sahani, Zhu, Cho, Machine learning-based intrusion detection for smart grid computing: a survey, ACM Trans Cyber-Phys Syst
Sani, Lei, Neagu, Computational complexity analysis of decision tree algorithms
Shahidi, Williams, Hassani, Physical activity during COVID-19 quarantine, Acta Paediatr
Singh, Ujjainiya, Prakash, A machine learningbased approach to determine infection status in recipients of BBV152 (Covaxin) whole-virion inactivated SARS-CoV-2 vaccine for serological surveys, Comput Biol Med
Stefan, Metabolic disorders, COVID-19 and vaccine-breakthrough infections, Nat Rev Endocrinol
Tegally, Moir, Everatt, Emergence of SARS-CoV-2 omicron lineages BA. 4 and BA. 5 in South Africa, Nat Med
Wang, Chen, Hozumi, Emerging vaccine-breakthrough SARS-CoV-2 variants, ACS Infect Dis
Wang, Li, Chiang, Machine learning prediction of COVID-19 severity levels from salivaomics data
Wedlund, Kvedar, New machine learning model predicts who may benefit most from COVID-19 vaccination, NPJ Digit Med
Woods, Hutchinson, Powers, The COVID-19 pandemic and physical activity, Sports Med Health Sci
Zoabi, Deri-Rozov, Shomron, Machine learningbased prediction of COVID-19 diagnosis based on symptoms, NPJ Digit Med
DOI record:
{
"DOI": "10.1177/20552076231207593",
"ISSN": [
"2055-2076",
"2055-2076"
],
"URL": "http://dx.doi.org/10.1177/20552076231207593",
"abstract": "<jats:sec><jats:title>Background</jats:title><jats:p> COVID-19 vaccines offer different levels of immune protection but do not provide 100% protection. Vaccinated persons with pre-existing comorbidities may be at an increased risk of SARS-CoV-2 breakthrough infection or reinfection. The aim of this study is to identify the critical variables associated with a higher probability of SARS-CoV-2 breakthrough infection using machine learning. </jats:p></jats:sec><jats:sec><jats:title>Methods</jats:title><jats:p> A dataset comprising symptoms and feedback from 257 persons, of whom 203 were vaccinated and 54 unvaccinated, was used for the investigation. Three machine learning algorithms – Deep Multilayer Perceptron (Deep MLP), XGBoost, and Logistic Regression – were trained with the original (imbalanced) dataset and the balanced dataset created by using the Random Oversampling Technique (ROT), and the Synthetic Minority Oversampling Technique (SMOTE). We compared the performance of the classification algorithms when the features highly correlated with breakthrough infection were used and when all features in the dataset were used. </jats:p></jats:sec><jats:sec><jats:title>Result</jats:title><jats:p> The results show that when highly correlated features were considered as predictors, with Random Oversampling to address data imbalance, the XGBoost classifier has the best performance (F1 = 0.96; accuracy = 0.96; AUC = 0.98; G-Mean = 0.98; MCC = 0.88). The Deep MLP had the second best performance (F1 = 0.94; accuracy = 0.94; AUC = 0.92; G-Mean = 0.70; MCC = 0.42), while Logistic Regression had less accurate performance (F1 = 0.89; accuracy = 0.88; AUC = 0.89; G-Mean = 0.89; MCC = 0.68). We also used Shapley Additive Explanations (SHAP) to investigate the interpretability of the models. We found that body temperature, total cholesterol, glucose level, blood pressure, waist circumference, body weight, body mass index (BMI), haemoglobin level, and physical activity per week are the most critical variables indicating a higher risk of breakthrough infection. </jats:p></jats:sec><jats:sec><jats:title>Conclusion</jats:title><jats:p> These results, evident from our unique data source derived from apparently healthy volunteers with cardiovascular risk factors, follow the expected pattern of positive or negative correlations previously reported in the literature. This information strengthens the body of knowledge currently applied in public health guidelines and may also be used by medical practitioners in the future to reduce the risk of SARS-CoV-2 breakthrough infection. </jats:p></jats:sec>",
"alternative-id": [
"10.1177/20552076231207593"
],
"author": [
{
"ORCID": "http://orcid.org/0000-0001-6340-078X",
"affiliation": [
{
"name": "Department of Information Technology, Faculty of Informatics and Design, Cape Peninsula University of Technology, Cape Town, South Africa"
}
],
"authenticated-orcid": false,
"family": "Daramola",
"given": "Olawande",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0002-9479-1143",
"affiliation": [
{
"name": "Department of Information Technology, Faculty of Informatics and Design, Cape Peninsula University of Technology, Cape Town, South Africa"
}
],
"authenticated-orcid": false,
"family": "Kavu",
"given": "Tatenda Duncan",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Division of Chemical Pathology, Department of Pathology, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa"
},
{
"name": "Division of Chemical Pathology, Department of Pathology, National Health Laboratory Service, Tygerberg Hospital, Cape Town, South Africa"
}
],
"family": "Kotze",
"given": "Maritha J",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Applied Microbial and Health Biotechnology Institute (AMHBI), Cape Peninsula University of Technology, Cape Town, South Africa"
},
{
"name": "Department of Biomedical Sciences, Faculty of Health and Wellness Sciences, Cape Peninsula University of Technology, Cape Town, South Africa"
}
],
"family": "Kamati",
"given": "Oiva",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Applied Microbial and Health Biotechnology Institute (AMHBI), Cape Peninsula University of Technology, Cape Town, South Africa"
}
],
"family": "Emjedi",
"given": "Zaakiyah",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Information Technology, Faculty of Informatics and Design, Cape Peninsula University of Technology, Cape Town, South Africa"
}
],
"family": "Kabaso",
"given": "Boniface",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-9220-649X",
"affiliation": [
{
"name": "St. Pölten University of Applied Sciences, St. Pölten, Austria"
}
],
"authenticated-orcid": false,
"family": "Moser",
"given": "Thomas",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5388-5952",
"affiliation": [
{
"name": "School of Health Information Science, University of Victoria, Victoria, BC, Canada"
}
],
"authenticated-orcid": false,
"family": "Stroetmann",
"given": "Karl",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Division of Epidemiology and Biostatistics, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa"
}
],
"family": "Fwemba",
"given": "Isaac",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Division of Epidemiology and Biostatistics, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa"
}
],
"family": "Daramola",
"given": "Fisayo",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Division of Epidemiology and Biostatistics, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa"
}
],
"family": "Nyirenda",
"given": "Martha",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Division of Chemical Pathology, Department of Pathology, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa"
}
],
"family": "van Rensburg",
"given": "Susan J",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Division of Epidemiology and Biostatistics, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa"
}
],
"family": "Nyasulu",
"given": "Peter S",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Applied Microbial and Health Biotechnology Institute (AMHBI), Cape Peninsula University of Technology, Cape Town, South Africa"
}
],
"family": "Marnewick",
"given": "Jeanine L",
"sequence": "additional"
}
],
"container-title": "DIGITAL HEALTH",
"container-title-short": "DIGITAL HEALTH",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"journals.sagepub.com"
]
},
"created": {
"date-parts": [
[
2023,
11,
6
]
],
"date-time": "2023-11-06T05:46:41Z",
"timestamp": 1699249601000
},
"deposited": {
"date-parts": [
[
2023,
11,
6
]
],
"date-time": "2023-11-06T05:46:48Z",
"timestamp": 1699249608000
},
"funder": [
{
"DOI": "10.13039/501100001322",
"award": [
"Data Analytics for Clinical Guidance on Immune Res"
],
"doi-asserted-by": "publisher",
"name": "South African Medical Research Council"
}
],
"indexed": {
"date-parts": [
[
2023,
11,
7
]
],
"date-time": "2023-11-07T00:56:23Z",
"timestamp": 1699318583891
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2023,
1
]
]
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "unspecified",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
1,
1
]
],
"date-time": "2023-01-01T00:00:00Z",
"timestamp": 1672531200000
}
}
],
"link": [
{
"URL": "http://journals.sagepub.com/doi/pdf/10.1177/20552076231207593",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "http://journals.sagepub.com/doi/full-xml/10.1177/20552076231207593",
"content-type": "application/xml",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "http://journals.sagepub.com/doi/pdf/10.1177/20552076231207593",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "179",
"original-title": [],
"prefix": "10.1177",
"published": {
"date-parts": [
[
2023,
1
]
]
},
"published-online": {
"date-parts": [
[
2023,
11,
5
]
]
},
"published-print": {
"date-parts": [
[
2023,
1
]
]
},
"publisher": "SAGE Publications",
"reference": [
{
"DOI": "10.1038/s41746-020-00372-6",
"doi-asserted-by": "publisher",
"key": "bibr1-20552076231207593"
},
{
"DOI": "10.1007/s40121-022-00600-4",
"doi-asserted-by": "publisher",
"key": "bibr2-20552076231207593"
},
{
"DOI": "10.3389/fmed.2022.948917",
"doi-asserted-by": "publisher",
"key": "bibr3-20552076231207593"
},
{
"DOI": "10.1016/j.compbiomed.2022.105419",
"doi-asserted-by": "publisher",
"key": "bibr4-20552076231207593"
},
{
"author": "Chen Y",
"journal-title": "SSRN Electron J",
"key": "bibr5-20552076231207593"
},
{
"DOI": "10.1021/acsinfecdis.1c00557",
"doi-asserted-by": "publisher",
"key": "bibr6-20552076231207593"
},
{
"DOI": "10.3389/fimmu.2022.811952",
"doi-asserted-by": "publisher",
"key": "bibr7-20552076231207593"
},
{
"DOI": "10.3390/informatics8040063",
"doi-asserted-by": "publisher",
"key": "bibr8-20552076231207593"
},
{
"DOI": "10.1142/9789813235533_0018",
"doi-asserted-by": "publisher",
"key": "bibr9-20552076231207593"
},
{
"DOI": "10.1145/2939672.2939785",
"doi-asserted-by": "publisher",
"key": "bibr10-20552076231207593"
},
{
"DOI": "10.1016/j.jclinepi.2020.03.002",
"doi-asserted-by": "publisher",
"key": "bibr11-20552076231207593"
},
{
"DOI": "10.1007/978-3-319-98074-4_2",
"doi-asserted-by": "publisher",
"key": "bibr12-20552076231207593"
},
{
"DOI": "10.1613/jair.953",
"doi-asserted-by": "publisher",
"key": "bibr13-20552076231207593"
},
{
"DOI": "10.1016/j.compbiomed.2022.105986",
"doi-asserted-by": "publisher",
"key": "bibr14-20552076231207593"
},
{
"key": "bibr15-20552076231207593",
"unstructured": "Wang A, Li F, Chiang S, et al. Machine learning prediction of COVID-19 severity levels from salivaomics data. ArXiv, https://www.ncbi.nlm.nih.gov/pubmed/35859658 (2022)."
},
{
"DOI": "10.1038/s41746-021-00425-4",
"doi-asserted-by": "publisher",
"key": "bibr16-20552076231207593"
},
{
"DOI": "10.1016/j.cmpb.2022.106981",
"doi-asserted-by": "publisher",
"key": "bibr17-20552076231207593"
},
{
"DOI": "10.1371/journal.pone.0280026",
"doi-asserted-by": "publisher",
"key": "bibr18-20552076231207593"
},
{
"DOI": "10.3390/healthcare11010031",
"doi-asserted-by": "publisher",
"key": "bibr19-20552076231207593"
},
{
"DOI": "10.1186/s12889-021-12383-3",
"doi-asserted-by": "publisher",
"key": "bibr20-20552076231207593"
},
{
"DOI": "10.1016/j.imu.2022.100908",
"doi-asserted-by": "publisher",
"key": "bibr21-20552076231207593"
},
{
"DOI": "10.3389/fcimb.2022.932204",
"doi-asserted-by": "publisher",
"key": "bibr22-20552076231207593"
},
{
"DOI": "10.1038/s41591-022-01911-2",
"doi-asserted-by": "publisher",
"key": "bibr23-20552076231207593"
},
{
"DOI": "10.1016/j.ijid.2022.11.024",
"doi-asserted-by": "publisher",
"key": "bibr24-20552076231207593"
},
{
"DOI": "10.1016/j.smhs.2020.05.006",
"doi-asserted-by": "publisher",
"key": "bibr25-20552076231207593"
},
{
"author": "Lundberg SM",
"first-page": "4768",
"key": "bibr26-20552076231207593",
"volume-title": "Proceedings of the 31st international conference on neural information processing systems",
"year": "2017"
},
{
"DOI": "10.1186/s12864-019-6413-7",
"doi-asserted-by": "publisher",
"key": "bibr27-20552076231207593"
},
{
"DOI": "10.1007/s42979-020-00394-7",
"doi-asserted-by": "publisher",
"key": "bibr28-20552076231207593"
},
{
"DOI": "10.1145/1412700.1412710",
"doi-asserted-by": "publisher",
"key": "bibr29-20552076231207593"
},
{
"author": "Sani HM",
"first-page": "191",
"key": "bibr30-20552076231207593",
"volume-title": "Artificial intelligence XXXV: 38th SGAI international conference on artificial intelligence, AI 2018"
},
{
"DOI": "10.1145/3578366",
"doi-asserted-by": "publisher",
"key": "bibr31-20552076231207593"
},
{
"author": "Kulkarni A",
"first-page": "1",
"journal-title": "ACM J Emerging Technol Comput Syst (JETC)",
"key": "bibr32-20552076231207593",
"volume": "13",
"year": "2016"
},
{
"author": "Kubat M",
"first-page": "179",
"journal-title": "Proc 14th Int Conf Machine Learn",
"key": "bibr33-20552076231207593",
"volume": "1",
"year": "1997"
},
{
"DOI": "10.1016/S1532-0464(03)00034-0",
"doi-asserted-by": "publisher",
"key": "bibr34-20552076231207593"
},
{
"DOI": "10.3390/jcm9061909",
"doi-asserted-by": "publisher",
"key": "bibr35-20552076231207593"
},
{
"DOI": "10.1038/s41574-021-00608-9",
"doi-asserted-by": "publisher",
"key": "bibr36-20552076231207593"
},
{
"DOI": "10.1016/j.numecd.2021.06.016",
"doi-asserted-by": "publisher",
"key": "bibr37-20552076231207593"
},
{
"DOI": "10.1016/j.bbalip.2020.158849",
"doi-asserted-by": "publisher",
"key": "bibr38-20552076231207593"
},
{
"DOI": "10.1016/S2213-8587(20)30152-2",
"doi-asserted-by": "publisher",
"key": "bibr39-20552076231207593"
},
{
"DOI": "10.3390/v14020336",
"doi-asserted-by": "publisher",
"key": "bibr40-20552076231207593"
},
{
"DOI": "10.3389/fimmu.2020.576818",
"doi-asserted-by": "publisher",
"key": "bibr41-20552076231207593"
},
{
"DOI": "10.1111/apa.15420",
"doi-asserted-by": "publisher",
"key": "bibr42-20552076231207593"
},
{
"author": "Kotze MJ",
"journal-title": "Clin Chem Lab Med",
"key": "bibr43-20552076231207593",
"volume": "61",
"year": "2023"
}
],
"reference-count": 43,
"references-count": 43,
"relation": {},
"resource": {
"primary": {
"URL": "http://journals.sagepub.com/doi/10.1177/20552076231207593"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Health Information Management",
"Computer Science Applications",
"Health Informatics",
"Health Policy"
],
"subtitle": [],
"title": "Detecting the most critical clinical variables of COVID-19 breakthrough infection in vaccinated persons using machine learning",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1177/sage-journals-update-policy",
"volume": "9"
}