Human protein interaction networks of ancestral and variant SARS-CoV-2 in organ-specific cells and bodily fluids
et al., Nature Communications, doi:10.1038/s41467-025-60949-1, Jul 2025
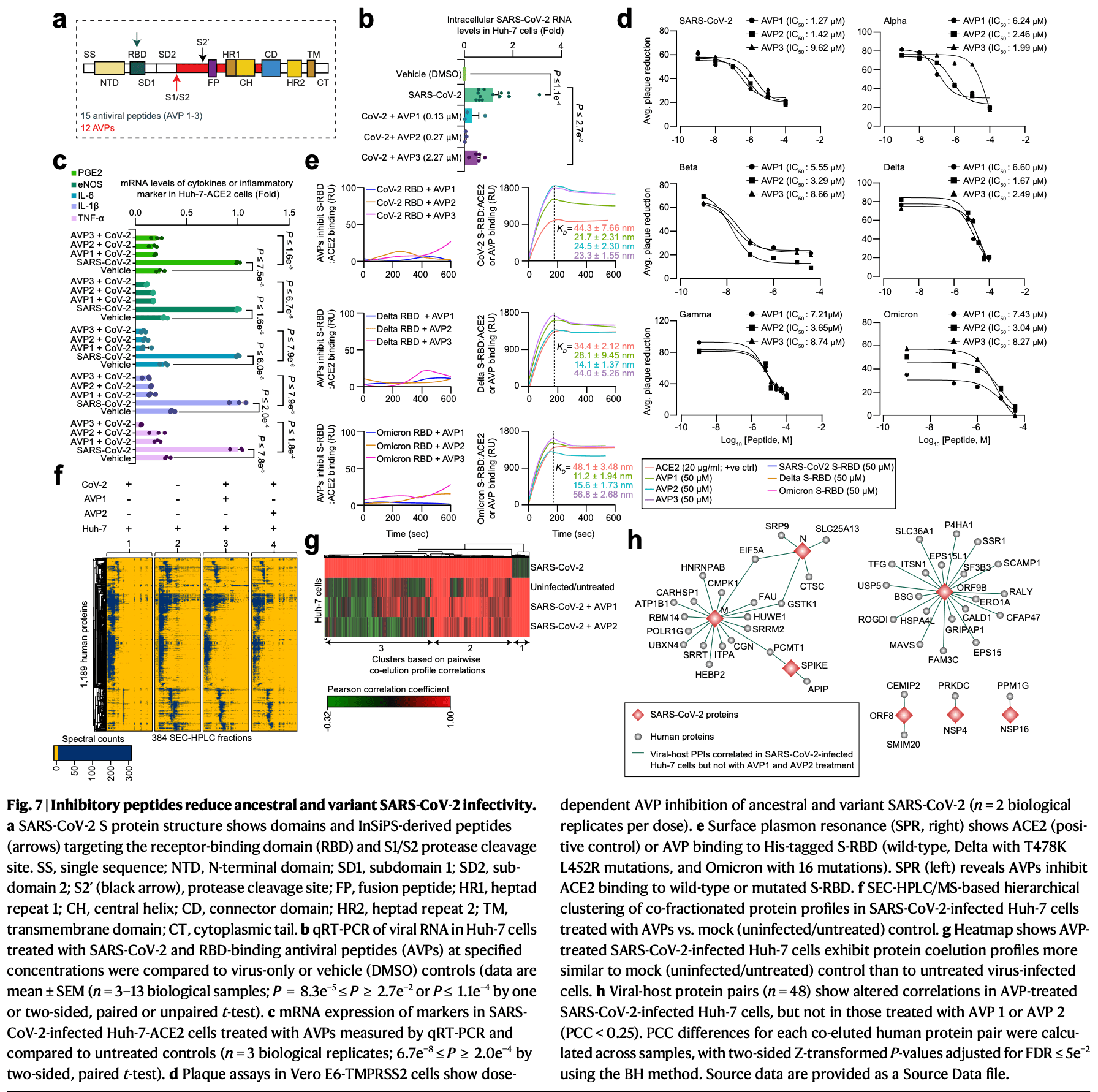
In vitro analysis showing synthetic peptides, fostamatinib, and NSP3-targeting strategies inhibit SARS-CoV-2 and variant replication in human liver, kidney, and lung cell lines. Authors conducted comprehensive mapping of SARS-CoV-2 and variant protein interactions with human proteins across five organ-derived cell lines and saliva from COVID-19 patients using affinity purification and co-fractionation mass spectrometry. The merged network identified 6,097 viral-host protein interactions, revealing previously unreported infection mechanisms including NSP3's interaction with fibrinogen that contributes to coagulation abnormalities, and NSP3's binding to IFIT5 to evade innate immunity.
Broderick et al., 1 Jul 2025, peer-reviewed, 29 authors.
Contact: mohan.babu@uregina.ca.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Human protein interaction networks of ancestral and variant SARS-CoV-2 in organ-specific cells and bodily fluids
Nature Communications, doi:10.1038/s41467-025-60949-1
Human protein interaction networks of ancestral and variant SARS-CoV-2 in organ-specific cells and bodily fluids A list of authors and their affiliations appears at the end of the paper Understanding SARS-CoV-2 human protein-protein interactions (PPIs) and the host response to infection is essential for developing effective COVID-19 antivirals. However, how the ancestral virus and its variants remodel virus-host protein assemblies in various organ-specific cells and bodily fluids remains unclear. Here, we conduct 639 affinity-purifications by tagging and expressing 28 SARS-CoV-2 and spike proteins from the ancestral virus and four variants in eight cell lines representing five mammalian organs and the immune system. Using mass spectrometry (MS), we identify both known and previously unreported SARS-CoV-2-human PPIs, highlighting similarities and differences across organ-or immune-derived cell lines and virus strains. Besides verifying the cell-and variant-specific PPIs, co-fractionation-MS analysis of COVID-19 patients' saliva confirm host PPI changes between SARS-CoV-2 strains. We discover that the NSP3 papain-like protease, a secreted protein, binds fibrinogen to induce abnormal blood clotting and interferon-induced proteins to evade host innate immune responses. Leveraging deep learning, we design peptide inhibitors that successfully blocked SARS-CoV-2 and variant replication in human liver cells, reversing virus-induced PPI alterations. Together, these findings provide molecular insights into SARS-CoV-2 biology, uncover reorganized viral-host protein assemblies during infection, and identify potential host therapeutic targets and inhibitors for developing antivirals against SARS-CoV-2 strains. The ancestral SARS-CoV-2 virus and its variants of concern (e.g., Alpha B. 1.1.7, Beta B.1.351, Gamma P.1, Delta B.1.617.2, Omicron B.1.1.529) or interest (e.g., Lambda C.37) causing coronavirus disease 2019 (COVID-19) have been detected in various organs (e.g., liver, lung, kidney) and bodily fluids (e.g., saliva) 1,2 , with organ-specific SARS-CoV-2 evolution observed in long COVID cases 3 . Studies using mass spectrometry (MS)-based proteomics and yeast-two hybrid (Y2H) assays highlight the roles for SARS-CoV-2 proteins in pathogenesis and host factors or processes targeted during infection [4] [5] [6] [7] [8] . Recent findings reveal that virus-host protein-protein interactions (PPIs) differ in variants, aiding immune evasion 9 . However, published studies focused on cell-based models or peripheral blood mononuclear cells, leaving gaps in understanding how SARS-CoV-2 and its variants remodel host responses and PPIs in various organs and fluids, particularly in saliva, a key site for SARS-CoV-2 infection and transmission 2 . While repurposed drugs and vaccines have reduced COVID-19 deaths, the need for improved therapeutics or organ-specific treatments continues. Targeting virus-host interface is a key strategy for antiviral development, with..
Reporting summary Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Author contributions
Competing interests The authors declare no competing interests.
Additional information Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41467-025-60949-1 . Correspondence and requests for materials should be addressed to Mohan Babu. Peer review information Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available. Reprints and permissions information is available at http://www.nature.com/reprints Publisher's note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Babu, Global landscape of cell envelope protein complexes in Escherichia coli, Nat. Biotechnol
Baggen, Vanstreels, Jansen, Daelemans, Cellular host factors for SARS-CoV-2 infection, Nat. Microbiol
Bojkova, Proteomics of SARS-CoV-2-infected host cells reveals therapy targets, Nature
Bouhaddou, SARS-CoV-2 variants evolve convergent strategies to remodel the host response, Cell
Broderick, Frank Dehne 9, Prasad, Joanne Lemieux 4, Cochrane 12 et al., Ali Hosseinnia 1 , Zoe Istace 1 , Maryam Hajikarimlou 3, Mohamed Taha Moutaoufik
Caillet-Saguy, Host PDZ-containing proteins targeted by SARS-CoV-2, FEBS J
Carabelli, SARS-CoV-2 variant biology: immune escape, transmission and fitness, Nat. Rev. Microbiol
Cione, Neuron-specific enolase serum levels in COVID-19 are related to the severity of lung injury, PLoS One
Colonna, Butovsky, Microglia function in the central nervous system during health and neurodegeneration, Annu. Rev. Immunol
Daniloski, Identification of required host factors for SARS-CoV-2 Infection in human cells, Cell
Davies, Plate, The glycoprotein quality control factor Malectin promotes coronavirus replication and viral protein biogenesis, eLife
Doolittle, Clotting of mammalian fibrinogens by papain: a reexamination, Biochemistry
Echavarria-Consuegra, Manipulation of the unfolded protein response: A pharmacological strategy against coronavirus infection, PLoS Pathog
Essa, Wu, Batumalaie, Sekar, Poh, Antiviral peptides against SARS-CoV-2: therapeutic targets, mechanistic antiviral activity, and efficient delivery, Pharm. Rep
Finkel, SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis, Nature
Garcia-Carceles, Caballero, Gil, Martinez, Kinase Inhibitors as Underexplored Antiviral Agents, J. Med Chem
Giannakopoulos, ISG15 Arg151 and the ISG15-conjugating enzyme UbE1L are important for innate immune control of Sindbis virus, J. Virol
Gordon, A SARS-CoV-2 protein interaction map reveals targets for drug repurposing, Nature
Gordon, Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms, Science
Grand, SARS-CoV-2 and the DNA damage response, J. Gen. Virol
Hajikarimlou, A computational approach to rapidly design peptides that detect SARS-CoV-2 surface protein S, NAR Genom. Bioinform
Han, SARS-CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG-I/MDA-5-MAVS, TLR3-TRIF, and cGAS-STING signaling pathways, J. Med Virol
Havugimana, A census of human soluble protein complexes, Cell
Huang, Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, Lancet
Huang, SARS-CoV-2 infection of the oral cavity and saliva, Nat. Med
Huttlin, The BioPlex Network: A Systematic Exploration of the Human Interactome, Cell
Kifle, Bruton tyrosine kinase inhibitors as potential therapeutic agents for COVID-19: A review, Metab. Open
Kim, A proteome-scale map of the SARS-CoV-2-human contactome, Nat. Biotechnol
Lai, Mice lacking the ISG15 E1 enzyme UbE1L demonstrate increased susceptibility to both mouse-adapted and nonmouse-adapted influenza B virus infection, J. Virol
Lan, Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor, Nature
Laurent, Global BioID-based SARS-CoV-2 proteins proximal interactome unveils novel ties between viral polypeptides and host factors involved in multiple COVID19-associated mechanisms, bioRxiv
Law, SARS-COV-2 recombinant Receptor-Binding-Domain (RBD) induces neutralizing antibodies against variant strains of SARS-CoV-2 and SARS-CoV-1, Vaccine
Li, Virus-host interactome and proteomic survey reveal potential virulence factors influencing SARS-CoV-2 Pathogenesis, Med
Liu, SARS-CoV-2-host proteome interactions for antiviral drug discovery, Mol. Syst. Biol
Malty, A map of human mitochondrial protein interactions linked to neurodegeneration reveals new mechanisms of redox homeostasis and NF-kappaB signaling, Cell Syst
Medved, Weisel, Fibrinogen & Factor, Os, Oi et al., Recommendations for nomenclature on fibrinogen and fibrin, J. Thromb. Haemost
Meidaninikjeh, Monocytes and macrophages in COVID-19: Friends and foes, Life Sci
Minkoff, Tenoever, Innate immune evasion strategies of SARS-CoV-2, Nat. Rev. Microbiol
Monje, Iwasaki, The neurobiology of long COVID, Neuron
Munnur, Altered ISGylation drives aberrant macrophagedependent immune responses during SARS-CoV-2 infection, Nat. Immunol
Nepusz, Yu, Paccanaro, Detecting overlapping protein complexes in protein-protein interaction networks, Nat. Methods
Scherer, SARS-CoV-2 nucleocapsid protein adheres to replication organelles before viral assembly at the Golgi/ERGIC and lysosome-mediated egress, Sci. Adv
Schindewolf, SARS-CoV-2 uses nonstructural Protein 16 to evade restriction by IFIT1 and IFIT3, J. Virol
Schrader, The clinically approved MEK inhibitor Trametinib efficiently blocks influenza A virus propagation and cytokine expression, Antivir. Res
Shin, Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity, Nature
Singh, Identification of multi-targeting natural antiviral peptides to impede SARS-CoV-2 infection, Struct. Chem
Stein, SARS-CoV-2 infection and persistence in the human body and brain at autopsy, Nature
Sui, Noubouossie, Gandotra, Cao, Elevated plasma fibrinogen is associated with excessive inflammation and disease severity in COVID-19 patients, Front. Cell Infect. Microbiol
Thul, A subcellular map of the human proteome, Science
Valikangas, COVID-19-specific transcriptomic signature detectable in blood across multiple cohorts, Front Genet
Van Cleemput, Organ-specific genome diversity of replication-competent SARS-CoV-2, Nat. Commun
Verschueren, Scoring large-scale affinity purification mass spectrometry datasets with MiST, Curr. Protoc. Bioinforma
Wang, A deep proteome and transcriptome abundance atlas of 29 healthy human tissues, Mol. Syst. Biol
Wang, Genetic Screens Identify Host Factors for SARS-CoV-2 and Common Cold Coronaviruses, Cell
Wu, Secreted ORF8 induces monocytic pro-inflammatory cytokines through NLRP3 pathways in patients with severe COVID-19, iScience
Xia, Peptide-based pan-CoV fusion inhibitors maintain high potency against SARS-CoV-2 Omicron variant, Cell Res
Xie, Inhibition of MEK signaling prevents SARS-CoV2induced lung damage and improves the survival of infected mice, J. Med. Virol
Xu, Compartmentalization-aided interaction screening reveals extensive high-order complexes within the SARS-CoV-2 proteome, Cell Rep
Yagci, Serin, Acicbe, Zeren, Odabasi, The relationship between serum erythropoietin, hepcidin, and haptoglobin levels with disease severity and other biochemical values in patients with COVID-19, Int J. Lab Hematol
Yan, Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2, Science
Zhang, The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Iota, Proc. Natl Acad. Sci. USA
Zhang, Zhang, Interferon-stimulated gene 15 and the protein ISGylation system, J. Interferon Cytokine Res
Zhao, Denison, Huibregtse, Gygi, Krug, Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways, Proc. Natl. Acad. Sci. USA
Zhou, A comprehensive SARS-CoV-2-human proteinprotein interactome reveals COVID-19 pathobiology and potential host therapeutic targets, Nat. Biotechnol
Zilocchi, Co-fractionation-mass spectrometry to characterize native mitochondrial protein assemblies in mammalian neurons and brain, Nat. Protoc
DOI record:
{
"DOI": "10.1038/s41467-025-60949-1",
"ISSN": [
"2041-1723"
],
"URL": "http://dx.doi.org/10.1038/s41467-025-60949-1",
"alternative-id": [
"60949"
],
"article-number": "5784",
"assertion": [
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Received",
"name": "received",
"order": 1,
"value": "16 May 2024"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "9 June 2025"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "First Online",
"name": "first_online",
"order": 3,
"value": "1 July 2025"
},
{
"group": {
"label": "Competing interests",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 1,
"value": "The authors declare no competing interests."
}
],
"author": [
{
"affiliation": [],
"family": "Broderick",
"given": "Kirsten",
"sequence": "first"
},
{
"affiliation": [],
"family": "Moutaoufik",
"given": "Mohamed Taha",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Saccon",
"given": "Tatiana",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Malty",
"given": "Ramy",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Amin",
"given": "Shahreen",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-6306-0551",
"affiliation": [],
"authenticated-orcid": false,
"family": "Phanse",
"given": "Sadhna",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Joseph",
"given": "Thomson Patrick",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-6138-7267",
"affiliation": [],
"authenticated-orcid": false,
"family": "Zilocchi",
"given": "Mara",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hosseinnia",
"given": "Ali",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0009-0007-8581-0596",
"affiliation": [],
"authenticated-orcid": false,
"family": "Istace",
"given": "Zoe",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-0762-3465",
"affiliation": [],
"authenticated-orcid": false,
"family": "Hajikarimlou",
"given": "Maryam",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-5000-761X",
"affiliation": [],
"authenticated-orcid": false,
"family": "Abrar",
"given": "Sakib",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0009-0005-3032-5601",
"affiliation": [],
"authenticated-orcid": false,
"family": "Fisher",
"given": "Jade",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Brassard",
"given": "Raelynn",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Perera",
"given": "Ranawaka",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Kumar",
"given": "Anil",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0009-0005-9143-086X",
"affiliation": [],
"authenticated-orcid": false,
"family": "Aoki",
"given": "Hiroyuki",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Rahmatbakhsh",
"given": "Matineh",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Jessulat",
"given": "Matthew",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-6371-7493",
"affiliation": [],
"authenticated-orcid": false,
"family": "Kobasa",
"given": "Darwyn",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Dehne",
"given": "Frank",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Prasad",
"given": "Bhanu",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Gagarinova",
"given": "Alla",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Joanne Lemieux",
"given": "M.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Cochrane",
"given": "Alan",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-1861-3441",
"affiliation": [],
"authenticated-orcid": false,
"family": "Houry",
"given": "Walid A.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-7022-1173",
"affiliation": [],
"authenticated-orcid": false,
"family": "Aly",
"given": "Khaled A.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Golshani",
"given": "Ashkan",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-4118-6406",
"affiliation": [],
"authenticated-orcid": false,
"family": "Babu",
"given": "Mohan",
"sequence": "additional"
}
],
"container-title": "Nature Communications",
"container-title-short": "Nat Commun",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"link.springer.com"
]
},
"created": {
"date-parts": [
[
2025,
7,
1
]
],
"date-time": "2025-07-01T12:27:54Z",
"timestamp": 1751372874000
},
"deposited": {
"date-parts": [
[
2025,
7,
1
]
],
"date-time": "2025-07-01T13:09:41Z",
"timestamp": 1751375381000
},
"funder": [
{
"DOI": "10.13039/501100000024",
"award": [
"COVID-19 SOF-549297-2019",
"VR3-172655",
"VS1-175520"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100000024",
"id-type": "DOI"
}
],
"name": "Gouvernement du Canada | Canadian Institutes of Health Research"
},
{
"DOI": "10.13039/501100000038",
"award": [
"DG-06009",
"DG-123456",
"DG-20234"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100000038",
"id-type": "DOI"
}
],
"name": "Gouvernement du Canada | Natural Sciences and Engineering Research Council of Canada"
},
{
"DOI": "10.13039/501100000196",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100000196",
"id-type": "DOI"
}
],
"name": "Canada Foundation for Innovation"
}
],
"indexed": {
"date-parts": [
[
2025,
7,
2
]
],
"date-time": "2025-07-02T04:13:27Z",
"timestamp": 1751429607071,
"version": "3.41.0"
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2025,
7,
1
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2025,
12
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by-nc-nd/4.0",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
7,
1
]
],
"date-time": "2025-07-01T00:00:00Z",
"timestamp": 1751328000000
}
},
{
"URL": "https://creativecommons.org/licenses/by-nc-nd/4.0",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
7,
1
]
],
"date-time": "2025-07-01T00:00:00Z",
"timestamp": 1751328000000
}
}
],
"link": [
{
"URL": "https://www.nature.com/articles/s41467-025-60949-1.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41467-025-60949-1",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41467-025-60949-1.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"prefix": "10.1038",
"published": {
"date-parts": [
[
2025,
7,
1
]
]
},
"published-online": {
"date-parts": [
[
2025,
7,
1
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference": [
{
"DOI": "10.1038/s41586-022-05542-y",
"author": "SR Stein",
"doi-asserted-by": "publisher",
"first-page": "758",
"journal-title": "Nature",
"key": "60949_CR1",
"unstructured": "Stein, S. R. et al. SARS-CoV-2 infection and persistence in the human body and brain at autopsy. Nature 612, 758–763 (2022).",
"volume": "612",
"year": "2022"
},
{
"DOI": "10.1038/s41591-021-01296-8",
"author": "N Huang",
"doi-asserted-by": "publisher",
"first-page": "892",
"journal-title": "Nat. Med.",
"key": "60949_CR2",
"unstructured": "Huang, N. et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat. Med. 27, 892–903 (2021).",
"volume": "27",
"year": "2021"
},
{
"DOI": "10.1038/s41467-021-26884-7",
"author": "J Van Cleemput",
"doi-asserted-by": "publisher",
"journal-title": "Nat. Commun.",
"key": "60949_CR3",
"unstructured": "Van Cleemput, J. et al. Organ-specific genome diversity of replication-competent SARS-CoV-2. Nat. Commun. 12, 6612 (2021).",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1038/s41587-022-01474-0",
"author": "Y Zhou",
"doi-asserted-by": "publisher",
"first-page": "128",
"journal-title": "Nat. Biotechnol.",
"key": "60949_CR4",
"unstructured": "Zhou, Y. et al. A comprehensive SARS-CoV-2-human protein-protein interactome reveals COVID-19 pathobiology and potential host therapeutic targets. Nat. Biotechnol. 41, 128–139 (2023).",
"volume": "41",
"year": "2023"
},
{
"DOI": "10.1038/s41586-020-2286-9",
"author": "DE Gordon",
"doi-asserted-by": "publisher",
"first-page": "459",
"journal-title": "Nature",
"key": "60949_CR5",
"unstructured": "Gordon, D. E. et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468 (2020).",
"volume": "583",
"year": "2020"
},
{
"DOI": "10.1038/s41587-022-01475-z",
"author": "DK Kim",
"doi-asserted-by": "publisher",
"first-page": "140",
"journal-title": "Nat. Biotechnol.",
"key": "60949_CR6",
"unstructured": "Kim, D. K. et al. A proteome-scale map of the SARS-CoV-2-human contactome. Nat. Biotechnol. 41, 140–149 (2023).",
"volume": "41",
"year": "2023"
},
{
"DOI": "10.15252/msb.202110396",
"author": "X Liu",
"doi-asserted-by": "publisher",
"journal-title": "Mol. Syst. Biol.",
"key": "60949_CR7",
"unstructured": "Liu, X. et al. SARS-CoV-2-host proteome interactions for antiviral drug discovery. Mol. Syst. Biol. 17, e10396 (2021).",
"volume": "17",
"year": "2021"
},
{
"DOI": "10.1016/j.medj.2020.07.002",
"author": "J Li",
"doi-asserted-by": "publisher",
"first-page": "99",
"journal-title": "Med",
"key": "60949_CR8",
"unstructured": "Li, J. et al. Virus-host interactome and proteomic survey reveal potential virulence factors influencing SARS-CoV-2 Pathogenesis. Med 2, 99–112.e117 (2021).",
"volume": "2",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2023.08.026",
"author": "M Bouhaddou",
"doi-asserted-by": "publisher",
"first-page": "4597",
"journal-title": "Cell",
"key": "60949_CR9",
"unstructured": "Bouhaddou, M. et al. SARS-CoV-2 variants evolve convergent strategies to remodel the host response. Cell 186, 4597–4614.e4526 (2023).",
"volume": "186",
"year": "2023"
},
{
"DOI": "10.1038/s41422-022-00617-x",
"author": "S Xia",
"doi-asserted-by": "publisher",
"first-page": "404",
"journal-title": "Cell Res.",
"key": "60949_CR10",
"unstructured": "Xia, S. et al. Peptide-based pan-CoV fusion inhibitors maintain high potency against SARS-CoV-2 Omicron variant. Cell Res. 32, 404–406 (2022).",
"volume": "32",
"year": "2022"
},
{
"DOI": "10.1093/nargab/lqac058",
"author": "M Hajikarimlou",
"doi-asserted-by": "publisher",
"journal-title": "NAR Genom. Bioinform.",
"key": "60949_CR11",
"unstructured": "Hajikarimlou, M. et al. A computational approach to rapidly design peptides that detect SARS-CoV-2 surface protein S. NAR Genom. Bioinform. 4, lqac058 (2022).",
"volume": "4",
"year": "2022"
},
{
"DOI": "10.1016/j.cels.2017.07.001",
"author": "RH Malty",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "Cell Syst.",
"key": "60949_CR12",
"unstructured": "Malty, R. H. et al. A map of human mitochondrial protein interactions linked to neurodegeneration reveals new mechanisms of redox homeostasis and NF-kappaB signaling. Cell Syst. 5, 1–14 (2017).",
"volume": "5",
"year": "2017"
},
{
"DOI": "10.1016/j.cell.2015.06.043",
"author": "EL Huttlin",
"doi-asserted-by": "publisher",
"first-page": "425",
"journal-title": "Cell",
"key": "60949_CR13",
"unstructured": "Huttlin, E. L. et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 162, 425–440 (2015).",
"volume": "162",
"year": "2015"
},
{
"DOI": "10.1002/0471250953.bi0819s49",
"author": "E Verschueren",
"doi-asserted-by": "publisher",
"first-page": "8 19 11",
"journal-title": "Curr. Protoc. Bioinforma.",
"key": "60949_CR14",
"unstructured": "Verschueren, E. et al. Scoring large-scale affinity purification mass spectrometry datasets with MiST. Curr. Protoc. Bioinforma. 49, 8 19 11–18 19 16 (2015).",
"volume": "49",
"year": "2015"
},
{
"key": "60949_CR15",
"unstructured": "Gordon, D. E. et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 370, eabe9403 (2020)."
},
{
"DOI": "10.1016/j.cell.2020.12.004",
"author": "R Wang",
"doi-asserted-by": "publisher",
"first-page": "106",
"journal-title": "Cell",
"key": "60949_CR16",
"unstructured": "Wang, R. et al. Genetic Screens Identify Host Factors for SARS-CoV-2 and Common Cold Coronaviruses. Cell 184, 106–119 e114 (2021).",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.10.030",
"author": "Z Daniloski",
"doi-asserted-by": "publisher",
"first-page": "92",
"journal-title": "Cell",
"key": "60949_CR17",
"unstructured": "Daniloski, Z. et al. Identification of required host factors for SARS-CoV-2 Infection in human cells. Cell 184, 92–105 e116 (2021).",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1038/s41564-021-00958-0",
"author": "J Baggen",
"doi-asserted-by": "publisher",
"first-page": "1219",
"journal-title": "Nat. Microbiol",
"key": "60949_CR18",
"unstructured": "Baggen, J., Vanstreels, E., Jansen, S. & Daelemans, D. Cellular host factors for SARS-CoV-2 infection. Nat. Microbiol 6, 1219–1232 (2021).",
"volume": "6",
"year": "2021"
},
{
"DOI": "10.1038/s41586-020-2332-7",
"author": "D Bojkova",
"doi-asserted-by": "publisher",
"first-page": "469",
"journal-title": "Nature",
"key": "60949_CR19",
"unstructured": "Bojkova, D. et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature 583, 469–472 (2020).",
"volume": "583",
"year": "2020"
},
{
"DOI": "10.15252/msb.20188503",
"author": "D Wang",
"doi-asserted-by": "publisher",
"journal-title": "Mol. Syst. Biol.",
"key": "60949_CR20",
"unstructured": "Wang, D. et al. A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol. Syst. Biol. 15, e8503 (2019).",
"volume": "15",
"year": "2019"
},
{
"DOI": "10.1126/sciadv.abl4895",
"author": "KM Scherer",
"doi-asserted-by": "publisher",
"journal-title": "Sci. Adv.",
"key": "60949_CR21",
"unstructured": "Scherer, K. M. et al. SARS-CoV-2 nucleocapsid protein adheres to replication organelles before viral assembly at the Golgi/ERGIC and lysosome-mediated egress. Sci. Adv. 8, eabl4895 (2022).",
"volume": "8",
"year": "2022"
},
{
"DOI": "10.1038/s41586-021-03610-3",
"author": "Y Finkel",
"doi-asserted-by": "publisher",
"first-page": "240",
"journal-title": "Nature",
"key": "60949_CR22",
"unstructured": "Finkel, Y. et al. SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis. Nature 594, 240–245 (2021).",
"volume": "594",
"year": "2021"
},
{
"DOI": "10.1371/journal.ppat.1009644",
"author": "L Echavarria-Consuegra",
"doi-asserted-by": "publisher",
"first-page": "e1009644",
"journal-title": "PLoS Pathog.",
"key": "60949_CR23",
"unstructured": "Echavarria-Consuegra, L. et al. Manipulation of the unfolded protein response: A pharmacological strategy against coronavirus infection. PLoS Pathog. 17, e1009644 (2021).",
"volume": "17",
"year": "2021"
},
{
"author": "AM Carabelli",
"first-page": "162",
"journal-title": "Nat. Rev. Microbiol",
"key": "60949_CR24",
"unstructured": "Carabelli, A. M. et al. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat. Rev. Microbiol 21, 162–177 (2023).",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.1371/journal.pone.0251819",
"author": "E Cione",
"doi-asserted-by": "publisher",
"first-page": "e0251819",
"journal-title": "PLoS One",
"key": "60949_CR25",
"unstructured": "Cione, E. et al. Neuron-specific enolase serum levels in COVID-19 are related to the severity of lung injury. PLoS One 16, e0251819 (2021).",
"volume": "16",
"year": "2021"
},
{
"DOI": "10.1111/febs.15881",
"author": "C Caillet-Saguy",
"doi-asserted-by": "publisher",
"first-page": "5148",
"journal-title": "FEBS J.",
"key": "60949_CR26",
"unstructured": "Caillet-Saguy, C. et al. Host PDZ-containing proteins targeted by SARS-CoV-2. FEBS J. 288, 5148–5162 (2021).",
"volume": "288",
"year": "2021"
},
{
"author": "JM Minkoff",
"first-page": "178",
"journal-title": "Nat. Rev. Microbiol",
"key": "60949_CR27",
"unstructured": "Minkoff, J. M. & tenOever, B. Innate immune evasion strategies of SARS-CoV-2. Nat. Rev. Microbiol 21, 178–194 (2023).",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.1016/j.cell.2012.08.011",
"author": "PC Havugimana",
"doi-asserted-by": "publisher",
"first-page": "1068",
"journal-title": "Cell",
"key": "60949_CR28",
"unstructured": "Havugimana, P. C. et al. A census of human soluble protein complexes. Cell 150, 1068–1081 (2012).",
"volume": "150",
"year": "2012"
},
{
"DOI": "10.1038/nbt.4024",
"author": "M Babu",
"doi-asserted-by": "publisher",
"first-page": "103",
"journal-title": "Nat. Biotechnol.",
"key": "60949_CR29",
"unstructured": "Babu, M. et al. Global landscape of cell envelope protein complexes in Escherichia coli. Nat. Biotechnol. 36, 103–112 (2018).",
"volume": "36",
"year": "2018"
},
{
"DOI": "10.1038/s41596-023-00901-z",
"doi-asserted-by": "crossref",
"key": "60949_CR30",
"unstructured": "Zilocchi, M. et al. Co-fractionation-mass spectrometry to characterize native mitochondrial protein assemblies in mammalian neurons and brain. Nat. Protoc. 18, 3918–3973 (2023)."
},
{
"DOI": "10.3389/fgene.2022.929887",
"author": "T Valikangas",
"doi-asserted-by": "publisher",
"journal-title": "Front Genet.",
"key": "60949_CR31",
"unstructured": "Valikangas, T. et al. COVID-19-specific transcriptomic signature detectable in blood across multiple cohorts. Front Genet. 13, 929887 (2022).",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1111/ijlh.13479",
"author": "S Yagci",
"doi-asserted-by": "publisher",
"first-page": "142",
"journal-title": "Int J. Lab Hematol.",
"key": "60949_CR32",
"unstructured": "Yagci, S., Serin, E., Acicbe, O., Zeren, M. I. & Odabasi, M. S. The relationship between serum erythropoietin, hepcidin, and haptoglobin levels with disease severity and other biochemical values in patients with COVID-19. Int J. Lab Hematol. 43, 142–151 (2021).",
"volume": "43",
"year": "2021"
},
{
"DOI": "10.1016/j.celrep.2021.109482",
"author": "W Xu",
"doi-asserted-by": "publisher",
"journal-title": "Cell Rep.",
"key": "60949_CR33",
"unstructured": "Xu, W. et al. Compartmentalization-aided interaction screening reveals extensive high-order complexes within the SARS-CoV-2 proteome. Cell Rep. 36, 109482 (2021).",
"volume": "36",
"year": "2021"
},
{
"DOI": "10.1146/annurev-immunol-051116-052358",
"author": "M Colonna",
"doi-asserted-by": "publisher",
"first-page": "441",
"journal-title": "Annu. Rev. Immunol.",
"key": "60949_CR34",
"unstructured": "Colonna, M. & Butovsky, O. Microglia function in the central nervous system during health and neurodegeneration. Annu. Rev. Immunol. 35, 441–468 (2017).",
"volume": "35",
"year": "2017"
},
{
"DOI": "10.1016/j.neuron.2022.10.006",
"author": "M Monje",
"doi-asserted-by": "publisher",
"first-page": "3484",
"journal-title": "Neuron",
"key": "60949_CR35",
"unstructured": "Monje, M. & Iwasaki, A. The neurobiology of long COVID. Neuron 110, 3484–3496 (2022).",
"volume": "110",
"year": "2022"
},
{
"DOI": "10.1016/j.lfs.2020.119010",
"author": "S Meidaninikjeh",
"doi-asserted-by": "publisher",
"journal-title": "Life Sci.",
"key": "60949_CR36",
"unstructured": "Meidaninikjeh, S. et al. Monocytes and macrophages in COVID-19: Friends and foes. Life Sci. 269, 119010 (2021).",
"volume": "269",
"year": "2021"
},
{
"DOI": "10.1016/j.metop.2021.100116",
"author": "ZD Kifle",
"doi-asserted-by": "publisher",
"journal-title": "Metab. Open",
"key": "60949_CR37",
"unstructured": "Kifle, Z. D. Bruton tyrosine kinase inhibitors as potential therapeutic agents for COVID-19: A review. Metab. Open 11, 100116 (2021).",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.1021/acs.jmedchem.1c00302",
"author": "J Garcia-Carceles",
"doi-asserted-by": "publisher",
"first-page": "935",
"journal-title": "J. Med Chem.",
"key": "60949_CR38",
"unstructured": "Garcia-Carceles, J., Caballero, E., Gil, C. & Martinez, A. Kinase Inhibitors as Underexplored Antiviral Agents. J. Med Chem. 65, 935–954 (2022).",
"volume": "65",
"year": "2022"
},
{
"DOI": "10.1002/jmv.28094",
"author": "J Xie",
"doi-asserted-by": "publisher",
"first-page": "6097",
"journal-title": "J. Med. Virol.",
"key": "60949_CR39",
"unstructured": "Xie, J. et al. Inhibition of MEK signaling prevents SARS-CoV2-induced lung damage and improves the survival of infected mice. J. Med. Virol. 94, 6097–6102 (2022).",
"volume": "94",
"year": "2022"
},
{
"DOI": "10.1016/j.antiviral.2018.07.006",
"author": "T Schrader",
"doi-asserted-by": "publisher",
"first-page": "80",
"journal-title": "Antivir. Res.",
"key": "60949_CR40",
"unstructured": "Schrader, T. et al. The clinically approved MEK inhibitor Trametinib efficiently blocks influenza A virus propagation and cytokine expression. Antivir. Res. 157, 80–92 (2018).",
"volume": "157",
"year": "2018"
},
{
"DOI": "10.1111/j.1538-7836.2008.03242.x",
"author": "L Medved",
"doi-asserted-by": "publisher",
"first-page": "355",
"journal-title": "J. Thromb. Haemost.",
"key": "60949_CR41",
"unstructured": "Medved, L., Weisel, J. W., Fibrinogen & Factor, X. S.oS. S. C.oI. S.oT. & Haemostasis. Recommendations for nomenclature on fibrinogen and fibrin. J. Thromb. Haemost. 7, 355–359 (2009).",
"volume": "7",
"year": "2009"
},
{
"DOI": "10.1016/j.isci.2023.106929",
"author": "X Wu",
"doi-asserted-by": "publisher",
"journal-title": "iScience",
"key": "60949_CR42",
"unstructured": "Wu, X. et al. Secreted ORF8 induces monocytic pro-inflammatory cytokines through NLRP3 pathways in patients with severe COVID-19. iScience 26, 106929 (2023).",
"volume": "26",
"year": "2023"
},
{
"DOI": "10.1021/bi5010987",
"author": "RF Doolittle",
"doi-asserted-by": "publisher",
"first-page": "6687",
"journal-title": "Biochemistry",
"key": "60949_CR43",
"unstructured": "Doolittle, R. F. Clotting of mammalian fibrinogens by papain: a re-examination. Biochemistry 53, 6687–6694 (2014).",
"volume": "53",
"year": "2014"
},
{
"DOI": "10.3389/fcimb.2021.734005",
"author": "J Sui",
"doi-asserted-by": "publisher",
"first-page": "734005",
"journal-title": "Front. Cell Infect. Microbiol.",
"key": "60949_CR44",
"unstructured": "Sui, J., Noubouossie, D. F., Gandotra, S. & Cao, L. Elevated plasma fibrinogen is associated with excessive inflammation and disease severity in COVID-19 patients. Front. Cell Infect. Microbiol. 11, 734005 (2021).",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.1016/S0140-6736(20)30183-5",
"author": "C Huang",
"doi-asserted-by": "publisher",
"first-page": "497",
"journal-title": "Lancet",
"key": "60949_CR45",
"unstructured": "Huang, C. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020).",
"volume": "395",
"year": "2020"
},
{
"DOI": "10.1101/2020.08.28.272955",
"doi-asserted-by": "crossref",
"key": "60949_CR46",
"unstructured": "Laurent, E. M. N. et al. Global BioID-based SARS-CoV-2 proteins proximal interactome unveils novel ties between viral polypeptides and host factors involved in multiple COVID19-associated mechanisms. bioRxiv, 2020.2008.2028. 272955 (2020)."
},
{
"DOI": "10.1038/s41590-021-01035-8",
"author": "D Munnur",
"doi-asserted-by": "publisher",
"first-page": "1416",
"journal-title": "Nat. Immunol.",
"key": "60949_CR47",
"unstructured": "Munnur, D. et al. Altered ISGylation drives aberrant macrophage-dependent immune responses during SARS-CoV-2 infection. Nat. Immunol. 22, 1416–1427 (2021).",
"volume": "22",
"year": "2021"
},
{
"DOI": "10.1128/jvi.01532-22",
"author": "C Schindewolf",
"doi-asserted-by": "publisher",
"journal-title": "J. Virol.",
"key": "60949_CR48",
"unstructured": "Schindewolf, C. et al. SARS-CoV-2 uses nonstructural Protein 16 to evade restriction by IFIT1 and IFIT3. J. Virol. 97, e0153222 (2023).",
"volume": "97",
"year": "2023"
},
{
"DOI": "10.1038/s41586-020-2601-5",
"author": "D Shin",
"doi-asserted-by": "publisher",
"first-page": "657",
"journal-title": "Nature",
"key": "60949_CR49",
"unstructured": "Shin, D. et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 587, 657–662 (2020).",
"volume": "587",
"year": "2020"
},
{
"DOI": "10.1073/pnas.0504754102",
"author": "C Zhao",
"doi-asserted-by": "publisher",
"first-page": "10200",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "60949_CR50",
"unstructured": "Zhao, C., Denison, C., Huibregtse, J. M., Gygi, S. & Krug, R. M. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl. Acad. Sci. USA 102, 10200–10205 (2005).",
"volume": "102",
"year": "2005"
},
{
"DOI": "10.1128/JVI.00105-08",
"author": "C Lai",
"doi-asserted-by": "publisher",
"first-page": "1147",
"journal-title": "J. Virol.",
"key": "60949_CR51",
"unstructured": "Lai, C. et al. Mice lacking the ISG15 E1 enzyme UbE1L demonstrate increased susceptibility to both mouse-adapted and non-mouse-adapted influenza B virus infection. J. Virol. 83, 1147–1151 (2009).",
"volume": "83",
"year": "2009"
},
{
"DOI": "10.1128/JVI.01590-08",
"author": "NV Giannakopoulos",
"doi-asserted-by": "publisher",
"first-page": "1602",
"journal-title": "J. Virol.",
"key": "60949_CR52",
"unstructured": "Giannakopoulos, N. V. et al. ISG15 Arg151 and the ISG15-conjugating enzyme UbE1L are important for innate immune control of Sindbis virus. J. Virol. 83, 1602–1610 (2009).",
"volume": "83",
"year": "2009"
},
{
"DOI": "10.1089/jir.2010.0110",
"author": "D Zhang",
"doi-asserted-by": "publisher",
"first-page": "119",
"journal-title": "J. Interferon Cytokine Res.",
"key": "60949_CR53",
"unstructured": "Zhang, D. & Zhang, D. E. Interferon-stimulated gene 15 and the protein ISGylation system. J. Interferon Cytokine Res. 31, 119–130 (2011).",
"volume": "31",
"year": "2011"
},
{
"DOI": "10.1007/s43440-022-00432-6",
"author": "RZ Essa",
"doi-asserted-by": "publisher",
"first-page": "1166",
"journal-title": "Pharm. Rep.",
"key": "60949_CR54",
"unstructured": "Essa, R. Z., Wu, Y. S., Batumalaie, K., Sekar, M. & Poh, C. L. Antiviral peptides against SARS-CoV-2: therapeutic targets, mechanistic antiviral activity, and efficient delivery. Pharm. Rep. 74, 1166–1181 (2022).",
"volume": "74",
"year": "2022"
},
{
"DOI": "10.1007/s11224-022-02113-9",
"doi-asserted-by": "crossref",
"key": "60949_CR55",
"unstructured": "Singh, S. et al. Identification of multi-targeting natural antiviral peptides to impede SARS-CoV-2 infection. Struct. Chem., 34, 1743–1758 (2023)."
},
{
"DOI": "10.1126/science.abb2762",
"author": "R Yan",
"doi-asserted-by": "publisher",
"first-page": "1444",
"journal-title": "Science",
"key": "60949_CR56",
"unstructured": "Yan, R. et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448 (2020).",
"volume": "367",
"year": "2020"
},
{
"DOI": "10.1002/jmv.27050",
"author": "L Han",
"doi-asserted-by": "publisher",
"first-page": "5376",
"journal-title": "J. Med Virol.",
"key": "60949_CR57",
"unstructured": "Han, L. et al. SARS-CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG-I/MDA-5-MAVS, TLR3-TRIF, and cGAS-STING signaling pathways. J. Med Virol. 93, 5376–5389 (2021).",
"volume": "93",
"year": "2021"
},
{
"DOI": "10.1073/pnas.2024202118",
"doi-asserted-by": "crossref",
"key": "60949_CR58",
"unstructured": "Zhang, Y. et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Iota. Proc. Natl Acad. Sci. USA 118, e2024202118 (2021)."
},
{
"DOI": "10.7554/eLife.100834",
"doi-asserted-by": "crossref",
"key": "60949_CR59",
"unstructured": "Davies, J. P. & Plate, L. The glycoprotein quality control factor Malectin promotes coronavirus replication and viral protein biogenesis. eLife 13, RP100834 (2024)."
},
{
"DOI": "10.1038/nmeth.1938",
"author": "T Nepusz",
"doi-asserted-by": "publisher",
"first-page": "471",
"journal-title": "Nat. Methods",
"key": "60949_CR60",
"unstructured": "Nepusz, T., Yu, H. & Paccanaro, A. Detecting overlapping protein complexes in protein-protein interaction networks. Nat. Methods 9, 471–472 (2012).",
"volume": "9",
"year": "2012"
},
{
"DOI": "10.1099/jgv.0.001918",
"doi-asserted-by": "crossref",
"key": "60949_CR61",
"unstructured": "Grand, R. J. SARS-CoV-2 and the DNA damage response. J. Gen. Virol. 104, 001918 (2023)."
},
{
"key": "60949_CR62",
"unstructured": "Thul, P. J. et al. A subcellular map of the human proteome. Science 356, eaal3321 (2017)."
},
{
"DOI": "10.1016/j.vaccine.2021.08.081",
"author": "JLM Law",
"doi-asserted-by": "publisher",
"first-page": "5769",
"journal-title": "Vaccine",
"key": "60949_CR63",
"unstructured": "Law, J. L. M. et al. SARS-COV-2 recombinant Receptor-Binding-Domain (RBD) induces neutralizing antibodies against variant strains of SARS-CoV-2 and SARS-CoV-1. Vaccine 39, 5769–5779 (2021).",
"volume": "39",
"year": "2021"
},
{
"DOI": "10.1038/s41586-020-2180-5",
"author": "J Lan",
"doi-asserted-by": "publisher",
"first-page": "215",
"journal-title": "Nature",
"key": "60949_CR64",
"unstructured": "Lan, J. et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020).",
"volume": "581",
"year": "2020"
}
],
"reference-count": 64,
"references-count": 64,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.nature.com/articles/s41467-025-60949-1"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Human protein interaction networks of ancestral and variant SARS-CoV-2 in organ-specific cells and bodily fluids",
"type": "journal-article",
"update-policy": "https://doi.org/10.1007/springer_crossmark_policy",
"volume": "16"
}