Differential abundance of lipids and metabolites related to SARS-CoV-2 infection and susceptibility
et al., Scientific Reports, doi:10.1038/s41598-023-40999-5, Sep 2023
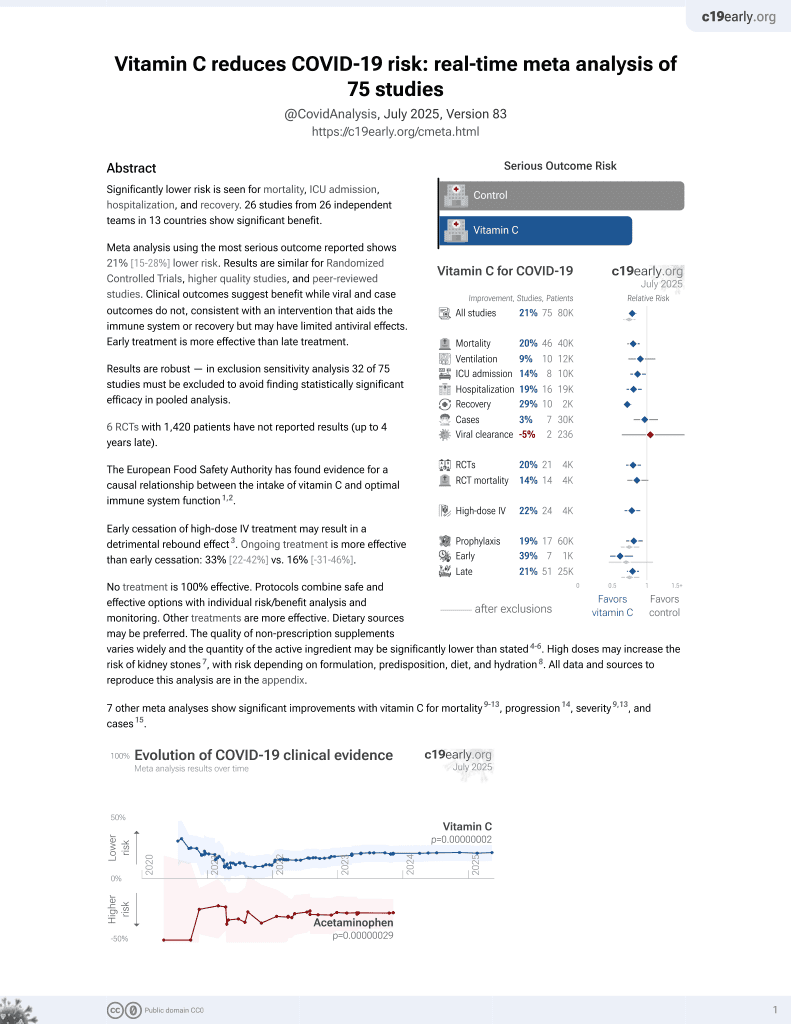
Vitamin C for COVID-19
6th treatment shown to reduce risk in
September 2020, now with p = 0.000000076 from 73 studies, recognized in 22 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
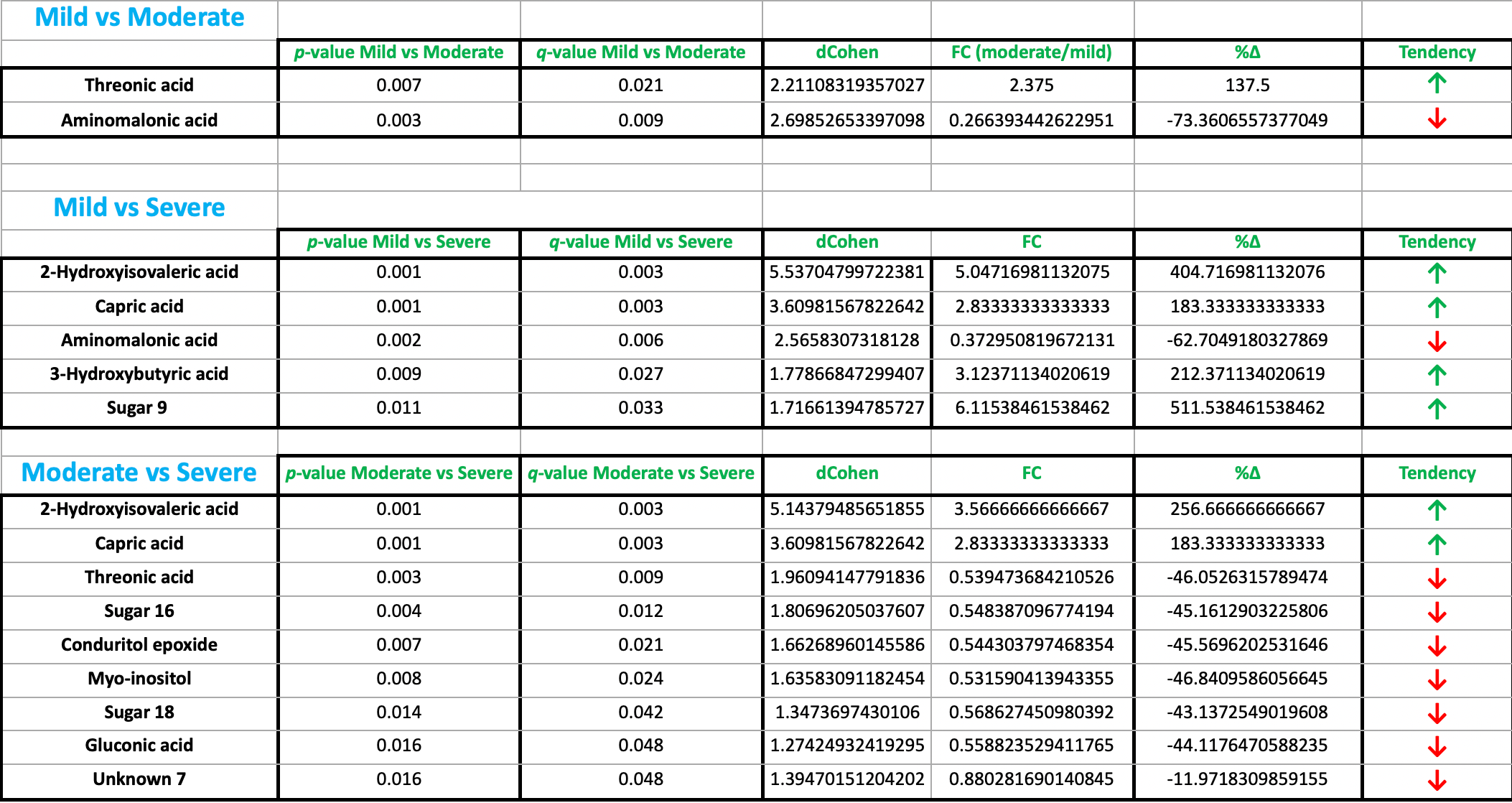
Plasma metabolomic analysis showing significantly lower threonic acid levels for severe and mild COVID-19 cases compared with moderate cases.
Threonic acid is a metabolite of vitamin C. The expected relationship in non-linear and depends on many factors, however higher vitamin C supports higher threonic acid levels. The results are consistent with a potential relationship between low vitamin C levels and COVID-19 severity:
In mild cases, low viral loads/inflammation may result in lower conversion of vitamin C to threonic acid.
In moderate cases, conversion of vitamin C to threonic acid may increase.
In severe COVID-19, extensive inflammation and oxidative stress may deplete vitamin C and threonic acid levels. Alternatively, low vitamin C levels may increase risk of COVID-19 severity. In either case, the results suggest that supplementation of vitamin C may be beneficial.
Albóniga et al., 13 Sep 2023, Spain, peer-reviewed, 13 authors.
Contact: sergio.serrano@salud.madrid.org, ceic.hrc@salud.madrid.org.
Differential abundance of lipids and metabolites related to SARS-CoV-2 infection and susceptibility
Scientific Reports, doi:10.1038/s41598-023-40999-5
The mechanisms driving SARS-CoV-2 susceptibility remain poorly understood, especially the factors determining why unvaccinated individuals remain uninfected despite high-risk exposures. To understand lipid and metabolite profiles related with COVID-19 susceptibility and disease progression. We collected samples from an exceptional group of unvaccinated healthcare workers heavily exposed to SARS-CoV-2 but not infected ('non-susceptible') and subjects who became infected during the follow-up ('susceptible'), including non-hospitalized and hospitalized patients with different disease severity providing samples at early disease stages. Then, we analyzed their plasma metabolomic profiles using mass spectrometry coupled with liquid and gas chromatography. We show specific lipids profiles and metabolites that could explain SARS-CoV-2 susceptibility and COVID-19 severity. More importantly, non-susceptible individuals show a unique lipidomic pattern characterized by the upregulation of most lipids, especially ceramides and sphingomyelin, which could be interpreted as markers of low susceptibility to SARS-CoV-2 infection. This study strengthens the findings of other researchers about the importance of studying lipid profiles as relevant markers of SARS-CoV-2 pathogenesis. Why some unvaccinated individuals with repeated high-risk exposures to SARS-CoV-2 did not show evidence of COVID-19 during the first pandemic waves? Among the possible explanations, host factors have been shown drive SARS-CoV-2 susceptibility and COVID-19 disease severity 1 . Consequently, omics studies, including metabolomic profiling, have gained attention to elucidate biochemical pathways affected by SARS-CoV-2 infection 2-4 . Metabolic profiles can be obtained by mass spectrometry (MS) coupled with different separation techniques, such as liquid chromatography (LC-MS), gas chromatography (GC-MS), or capillary electrophoresis (CE-MS). These techniques allow the identification of different types of molecules, such as amino acids, lipids, and glycoproteins 5 . Since it is not possible to analyze the vast amount of metabolites present in plasma samples using only one technique, the combination of these complementary techniques provides a broader picture of the metabolic pathways and their metabolites under selected conditions. The COVID-19 pandemic has generated a considerable economic and societal impact 6 including long-term effects 7,8 due to Long COVID conditions 9 . Abnormal immune responses to SARS-CoV-2 characterized by impaired macrophage, neutrophil, and dendritic functions or decreased IFN-ɣ production appear to explain adverse clinical outcomes [10] [11] [12] . Furthermore, several comorbidities such as immunosuppression, diabetes, pulmonary disease, or cardiovascular disease negatively affect disease progression 13, 14 . Lipidomics and metabolomics
Competing interests The authors declare no competing interests.
Additional information
Supplementary Information The online version contains supplementary material available at https:// doi. org/ 10. 1038/ s41598-023-40999-5. Correspondence and requests for materials should be addressed to S.S.-V. Reprints and permissions information is available at www.nature.com/reprints. Publisher's note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Aguilar, Current understanding of COVID-19 clinical course and investigational treatments, Front. Med
Albeituni, Stiban, Roles of ceramides and other sphingolipids in immune cell function and inflammation
Albóniga, Metabolic snapshot of plasma samples reveals new pathways implicated in SARS-CoV-2 pathogenesis, J. Proteome Res
Alexander, Acute kidney injury in severe COVID-19 has similarities to sepsis-associated kidney injury, Mayo Clin. Proc
Attaway, Scheraga, Bhimraj, Biehl, Hatipoğlu, Severe covid-19 pneumonia: pathogenesis and clinical management, BMJ, doi:10.1136/bmj.n436
Barberis, Large-scale plasma analysis revealed new mechanisms and molecules associated with the host response to SARS-CoV-2, Int. J. Mol. Sci
Barberis, Large-scale plasma analysis revealed new mechanisms and molecules associated with the host response to sarscov-2, Int. J. Mol. Sci
Bizkarguenaga, Uneven metabolic and lipidomic profiles in recovered COVID-19 patients as investigated by plasma NMR metabolomics, NMR Biomed
Bruzzone, Conde, Embade, Mato, Millet, Metabolomics as a powerful tool for diagnostic, pronostic and drug intervention analysis in COVID-19, Front. Mol. Biosci
Byeon, Development of a multiomics model for identification of predictive biomarkers for COVID-19 severity: A retrospective cohort study, Lancet Digit. Health
Casari, Manfredi, Metharom, Falasca, Dissecting lipid metabolism alterations in SARS-CoV-2, Prog. Lipid Res
Caterino, Dysregulation of lipid metabolism and pathological inflammation in patients with COVID-19, Sci. Rep
Chen, Blood molecular markers associated with COVID-19 immunopathology and multi-organ damage, EMBO J
Chen, Global prevalence of post-coronavirus disease 2019 (COVID-19) condition or long COVID: A meta-analysis and systematic review, J. Infect. Dis
Chen, Wang, Kuo, Tsai, Metabolome analysis for investigating host-gut microbiota interactions, J. Formos. Med. Assoc
Cobb, α-Hydroxybutyric acid is a selective metabolite biomarker of impaired glucose tolerance, Diabetes Care
Danlos, Metabolomic analyses of COVID-19 patients unravel stage-dependent and prognostic biomarkers, Cell Death Dis
Davis, Mccorkell, Vogel, Topol, Long, Major findings, mechanisms and recommendations, Nat. Rev. Microbiol
Dei Cas, Link between serum lipid signature and prognostic factors in COVID-19 patients, Sci. Rep
Di Gennaro, Long covid: A systematic review and meta-analysis of 120,970 patients, SSRN Electron. J, doi:10.2139/ssrn.4099429
Essalmani, Distinctive roles of furin and TMPRSS2 in SARS-CoV-2 infectivity, J. Virol
Fanelli, Acute respiratory distress syndrome: New definition, current and future therapeutic options, J. Thorac. Dis
Giamarellos-Bourboulis, Complex immune dysregulation in COVID-19 patients with severe respiratory failure, Cell Host Microbe
Gil-De-La-Fuente, CEU mass mediator 3.0: A metabolite annotation tool, J. Proteome Res
Gomez-Gomez, Untargeted detection of the carbonyl metabolome by chemical derivatization and liquid chromatographytandem mass spectrometry in precursor ion scan mode: Elucidation of COVID-19 severity biomarkers, Anal. Chim. Acta
Gordon, A SARS-CoV-2 protein interaction map reveals targets for drug repurposing, Nature
Gray, Diagnostic potential of the plasma lipidome in infectious disease: Application to acute SARS-CoV-2 infection, Metabolites
Gómez-Muñoz, Ceramide-1-phosphate promotes cell survival through activation of the phosphatidylinositol 3-kinase/ protein kinase B pathway, FEBS Lett
Hasan, Suleiman, Pérez-López, Metabolomics in the diagnosis and prognosis of COVID-19, Front. Genet
Hierholzer, Kabara, In vitro effects of monolaurin compounds on enveloped RNA and DNA viruses, J. Food Saf
Hmdb, Human Metabolome Database: Showing Metabocard for Threonic acid (HMDB0000943
Hmdb, Human Metabolome Database: Showing metabocard for Aminomalonic acid (HMDB0001147
Hou, n-3 polyunsaturated fatty acids suppress CD4+ T cell proliferation by altering phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2] organization, Biochim. Biophys. Acta BBA Biomembr
Johnson, Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis, Nature
Khodadoust, Inferring a causal relationship between ceramide levels and COVID-19 respiratory distress, Sci. Rep
Koelmel, Lipid annotator: Towards accurate annotation in non-targeted liquid chromatography high-resolution tandem mass spectrometry (LC-HRMS/MS) lipidomics using a rapid and user-friendly software, Metabolites
Kornhuber, Hoertel, Gulbins, The acid sphingomyelinase/ceramide system in COVID-19, Mol. Psychiatry
Kočar, Režen, Rozman, Cholesterol, lipoproteins, and COVID-19: Basic concepts and clinical applications, Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids
Kuligowski, Sánchez-Illana, Sanjuán-Herráez, Vento, Quintás, Intra-batch effect correction in liquid chromatography-mass spectrometry using quality control samples and support vector regression (QC-SVRC), Analyst
Kumar, Kumar, Arya, Anand, Priyadarshi, Association of COVID-19 with hepatic metabolic dysfunction, World J. Virol
Kurano, Dynamic modulations of sphingolipids and glycerophospholipids in COVID-19, Clin. Transl. Med
Köhler, Rose, Falk, Pauling, Investigating global lipidome alterations with the lipid network explorer, Metabolites
Lejardi, Technologies, Improving Coverage of the Plasma Lipidome Using Iterative MS/MS Data Acquisition Combined with Lipid Annotator Software and 6546
Ma, Long-term consequences of COVID-19 at 6 months and above: A systematic review and meta-analysis, Int. J. Environ. Res. Public. Health
Merad, Blish, Sallusto, Iwasaki, The immunology and immunopathology of COVID-19, Science
Milani, Macchi, Guz-Mark, Vitamin C in the treatment of COVID-19, Nutrients
Petrache, Marked elevations in lung and plasma ceramide in COVID-19 linked to microvascular injury, JCI Insight
Páez-Franco, Metabolomics analysis reveals a modified amino acid metabolism that correlates with altered oxygen homeostasis in COVID-19 patients, Sci. Rep
Ren, Alterations in the human oral and gut microbiomes and lipidomics in COVID-19, Gut
Richards, Economic burden of COVID-19: A systematic review, Clin. Outcomes Res
Schmelter, Metabolic and lipidomic markers differentiate COVID-19 from non-hospitalized and other intensive care patients, Front. Mol. Biosci
Shakaib, A comprehensive review on clinical and mechanistic pathophysiological aspects of COVID-19 Malady: How far have we come?, Virol. J
Shi, The serum metabolome of COVID-19 patients is distinctive and predictive, Metabolism
Sindelar, Longitudinal metabolomics of human plasma reveals prognostic markers of COVID-19 disease severity, Cell Rep. Med
Song, Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis, Cell Metab
Suzuki, Suzuki, Virus infection and lipid rafts, Biol. Pharm. Bull
Torres-Ruiz, Redefining COVID-19 severity and prognosis: The role of clinical and immunobiotypes, Front. Immunol
Torretta, Severity of COVID-19 patients predicted by serum sphingolipids signature, Int. J. Mol. Sci
Toscano, De Araújo, De Souza, Barbosa Mirabal, De Vasconcelos Torres, Vitamin C and D supplementation and the severity of COVID-19: A protocol for systematic review and meta-analysis, Medicine
Tsugawa, A lipidome atlas in MS-DIAL 4, Nat. Biotechnol
Törnquist, Asghar, Srinivasan, Korhonen, Lindholm, Sphingolipids as modulators of SARS-CoV-2 infection, Front. Cell Dev. Biol
Valdés, Metabolomics study of COVID-19 patients in four different clinical stages, Sci. Rep
Vardhana, Wolchok, The many faces of the anti-COVID immune response, J. Exp. Med
Wong, Damania, SARS-CoV-2 dependence on host pathways, Science
Wu, Plasma metabolomic and lipidomic alterations associated with COVID-19, Natl. Sci. Rev
Žarković, Lipidomics revealed plasma phospholipid profile differences between deceased and recovered COVID-19 patients, Biomolecules
DOI record:
{
"DOI": "10.1038/s41598-023-40999-5",
"ISSN": [
"2045-2322"
],
"URL": "http://dx.doi.org/10.1038/s41598-023-40999-5",
"abstract": "<jats:title>Abstract</jats:title><jats:p>The mechanisms driving SARS-CoV-2 susceptibility remain poorly understood, especially the factors determining why unvaccinated individuals remain uninfected despite high-risk exposures. To understand lipid and metabolite profiles related with COVID-19 susceptibility and disease progression. We collected samples from an exceptional group of unvaccinated healthcare workers heavily exposed to SARS-CoV-2 but not infected (‘non-susceptible’) and subjects who became infected during the follow-up (‘susceptible’), including non-hospitalized and hospitalized patients with different disease severity providing samples at early disease stages. Then, we analyzed their plasma metabolomic profiles using mass spectrometry coupled with liquid and gas chromatography. We show specific lipids profiles and metabolites that could explain SARS-CoV-2 susceptibility and COVID-19 severity. More importantly, non-susceptible individuals show a unique lipidomic pattern characterized by the upregulation of most lipids, especially ceramides and sphingomyelin, which could be interpreted as markers of low susceptibility to SARS-CoV-2 infection. This study strengthens the findings of other researchers about the importance of studying lipid profiles as relevant markers of SARS-CoV-2 pathogenesis.</jats:p>",
"alternative-id": [
"40999"
],
"article-number": "15124",
"assertion": [
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Received",
"name": "received",
"order": 1,
"value": "10 May 2023"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "20 August 2023"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "First Online",
"name": "first_online",
"order": 3,
"value": "13 September 2023"
},
{
"group": {
"label": "Competing interests",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 1,
"value": "The authors declare no competing interests."
}
],
"author": [
{
"affiliation": [],
"family": "Albóniga",
"given": "Oihane E.",
"sequence": "first"
},
{
"affiliation": [],
"family": "Moreno",
"given": "Elena",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Martínez-Sanz",
"given": "Javier",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Vizcarra",
"given": "Pilar",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ron",
"given": "Raquel",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Díaz-Álvarez",
"given": "Jorge",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Rosas",
"given": "Marta",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Sánchez-Conde",
"given": "Matilde",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Galán",
"given": "Juan Carlos",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Angulo",
"given": "Santiago",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Moreno",
"given": "Santiago",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Barbas",
"given": "Coral",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Serrano-Villar",
"given": "Sergio",
"sequence": "additional"
}
],
"container-title": "Scientific Reports",
"container-title-short": "Sci Rep",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"link.springer.com"
]
},
"created": {
"date-parts": [
[
2023,
9,
13
]
],
"date-time": "2023-09-13T09:02:15Z",
"timestamp": 1694595735000
},
"deposited": {
"date-parts": [
[
2023,
9,
13
]
],
"date-time": "2023-09-13T10:10:42Z",
"timestamp": 1694599842000
},
"funder": [
{
"DOI": "10.13039/100009947",
"award": [
"MISP# IIS 60257"
],
"doi-asserted-by": "publisher",
"name": "Merck Sharp and Dohme"
},
{
"DOI": "10.13039/501100004587",
"award": [
"AC17/00019, PI18/00154, COV20/00349, ICI20/00058, PI21/00141"
],
"doi-asserted-by": "publisher",
"name": "Instituto de Salud Carlos III"
},
{
"DOI": "10.13039/501100015756",
"award": [
"Mobility grant"
],
"doi-asserted-by": "publisher",
"name": "Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica"
}
],
"indexed": {
"date-parts": [
[
2023,
9,
14
]
],
"date-time": "2023-09-14T04:54:41Z",
"timestamp": 1694667281067
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2023,
9,
13
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2023,
12
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
9,
13
]
],
"date-time": "2023-09-13T00:00:00Z",
"timestamp": 1694563200000
}
},
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
9,
13
]
],
"date-time": "2023-09-13T00:00:00Z",
"timestamp": 1694563200000
}
}
],
"link": [
{
"URL": "https://www.nature.com/articles/s41598-023-40999-5.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41598-023-40999-5",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41598-023-40999-5.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"prefix": "10.1038",
"published": {
"date-parts": [
[
2023,
9,
13
]
]
},
"published-online": {
"date-parts": [
[
2023,
9,
13
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference": [
{
"DOI": "10.1126/science.abg6837",
"author": "JP Wong",
"doi-asserted-by": "publisher",
"first-page": "884",
"journal-title": "Science",
"key": "40999_CR1",
"unstructured": "Wong, J. P. & Damania, B. SARS-CoV-2 dependence on host pathways. Science 371, 884–885 (2021).",
"volume": "371",
"year": "2021"
},
{
"DOI": "10.1038/s41586-020-2286-9",
"author": "DE Gordon",
"doi-asserted-by": "publisher",
"first-page": "459",
"journal-title": "Nature",
"key": "40999_CR2",
"unstructured": "Gordon, D. E. et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468 (2020).",
"volume": "583",
"year": "2020"
},
{
"DOI": "10.15252/embj.2020105896",
"author": "Y Chen",
"doi-asserted-by": "publisher",
"first-page": "24",
"journal-title": "EMBO J.",
"key": "40999_CR3",
"unstructured": "Chen, Y. et al. Blood molecular markers associated with COVID-19 immunopathology and multi-organ damage. EMBO J. 39, 24 (2020).",
"volume": "39",
"year": "2020"
},
{
"DOI": "10.3390/metabo11070467",
"author": "N Gray",
"doi-asserted-by": "publisher",
"first-page": "467",
"journal-title": "Metabolites",
"key": "40999_CR4",
"unstructured": "Gray, N. et al. Diagnostic potential of the plasma lipidome in infectious disease: Application to acute SARS-CoV-2 infection. Metabolites 11, 467 (2021).",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.1016/j.jfma.2018.09.007",
"author": "MX Chen",
"doi-asserted-by": "publisher",
"first-page": "S10",
"journal-title": "J. Formos. Med. Assoc.",
"key": "40999_CR5",
"unstructured": "Chen, M. X., Wang, S.-Y., Kuo, C.-H. & Tsai, I.-L. Metabolome analysis for investigating host-gut microbiota interactions. J. Formos. Med. Assoc. 118, S10–S22 (2019).",
"volume": "118",
"year": "2019"
},
{
"DOI": "10.2147/CEOR.S338225",
"author": "F Richards",
"doi-asserted-by": "publisher",
"first-page": "293",
"journal-title": "Clin. Outcomes Res.",
"key": "40999_CR6",
"unstructured": "Richards, F. et al. Economic burden of COVID-19: A systematic review. Clin. Outcomes Res. 14, 293–307 (2022).",
"volume": "14",
"year": "2022"
},
{
"DOI": "10.3390/ijerph19116865",
"author": "Y Ma",
"doi-asserted-by": "publisher",
"first-page": "6865",
"journal-title": "Int. J. Environ. Res. Public. Health",
"key": "40999_CR7",
"unstructured": "Ma, Y. et al. Long-term consequences of COVID-19 at 6 months and above: A systematic review and meta-analysis. Int. J. Environ. Res. Public. Health 19, 6865 (2022).",
"volume": "19",
"year": "2022"
},
{
"DOI": "10.1093/infdis/jiac136",
"author": "C Chen",
"doi-asserted-by": "publisher",
"first-page": "1593",
"journal-title": "J. Infect. Dis.",
"key": "40999_CR8",
"unstructured": "Chen, C. et al. Global prevalence of post-coronavirus disease 2019 (COVID-19) condition or long COVID: A meta-analysis and systematic review. J. Infect. Dis. 226, 1593–1607 (2022).",
"volume": "226",
"year": "2022"
},
{
"DOI": "10.2139/ssrn.4099429",
"author": "F Di Gennaro",
"doi-asserted-by": "publisher",
"journal-title": "SSRN Electron. J.",
"key": "40999_CR9",
"unstructured": "Di Gennaro, F. et al. Long covid: A systematic review and meta-analysis of 120,970 patients. SSRN Electron. J. https://doi.org/10.2139/ssrn.4099429 (2022).",
"year": "2022"
},
{
"DOI": "10.1016/j.chom.2020.04.009",
"author": "EJ Giamarellos-Bourboulis",
"doi-asserted-by": "publisher",
"first-page": "992",
"journal-title": "Cell Host Microbe",
"key": "40999_CR10",
"unstructured": "Giamarellos-Bourboulis, E. J. et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe 27, 992-1000.e3 (2020).",
"volume": "27",
"year": "2020"
},
{
"DOI": "10.1084/jem.20200678",
"author": "SA Vardhana",
"doi-asserted-by": "publisher",
"journal-title": "J. Exp. Med.",
"key": "40999_CR11",
"unstructured": "Vardhana, S. A. & Wolchok, J. D. The many faces of the anti-COVID immune response. J. Exp. Med. 217, e20200678 (2020).",
"volume": "217",
"year": "2020"
},
{
"DOI": "10.1126/science.abm8108",
"author": "M Merad",
"doi-asserted-by": "publisher",
"first-page": "1122",
"journal-title": "Science",
"key": "40999_CR12",
"unstructured": "Merad, M., Blish, C. A., Sallusto, F. & Iwasaki, A. The immunology and immunopathology of COVID-19. Science 375, 1122–1127 (2022).",
"volume": "375",
"year": "2022"
},
{
"DOI": "10.1136/bmj.n436",
"author": "AH Attaway",
"doi-asserted-by": "publisher",
"journal-title": "BMJ",
"key": "40999_CR13",
"unstructured": "Attaway, A. H., Scheraga, R. G., Bhimraj, A., Biehl, M. & Hatipoğlu, U. Severe covid-19 pneumonia: pathogenesis and clinical management. BMJ https://doi.org/10.1136/bmj.n436 (2021).",
"year": "2021"
},
{
"DOI": "10.1186/s12985-021-01578-0",
"author": "B Shakaib",
"doi-asserted-by": "publisher",
"first-page": "120",
"journal-title": "Virol. J.",
"key": "40999_CR14",
"unstructured": "Shakaib, B. et al. A comprehensive review on clinical and mechanistic pathophysiological aspects of COVID-19 Malady: How far have we come?. Virol. J. 18, 120 (2021).",
"volume": "18",
"year": "2021"
},
{
"DOI": "10.3389/fmolb.2023.1111482",
"author": "C Bruzzone",
"doi-asserted-by": "publisher",
"first-page": "1111482",
"journal-title": "Front. Mol. Biosci.",
"key": "40999_CR15",
"unstructured": "Bruzzone, C., Conde, R., Embade, N., Mato, J. M. & Millet, O. Metabolomics as a powerful tool for diagnostic, pronostic and drug intervention analysis in COVID-19. Front. Mol. Biosci. 10, 1111482 (2023).",
"volume": "10",
"year": "2023"
},
{
"DOI": "10.1038/s41419-021-03540-y",
"author": "F-X Danlos",
"doi-asserted-by": "publisher",
"first-page": "258",
"journal-title": "Cell Death Dis.",
"key": "40999_CR16",
"unstructured": "Danlos, F.-X. et al. Metabolomic analyses of COVID-19 patients unravel stage-dependent and prognostic biomarkers. Cell Death Dis. 12, 258 (2021).",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1093/nsr/nwaa086",
"author": "D Wu",
"doi-asserted-by": "publisher",
"first-page": "1157",
"journal-title": "Natl. Sci. Rev.",
"key": "40999_CR17",
"unstructured": "Wu, D. et al. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl. Sci. Rev. 7, 1157–1168 (2020).",
"volume": "7",
"year": "2020"
},
{
"DOI": "10.1038/s41598-021-82426-7",
"author": "M Caterino",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Sci. Rep.",
"key": "40999_CR18",
"unstructured": "Caterino, M. et al. Dysregulation of lipid metabolism and pathological inflammation in patients with COVID-19. Sci. Rep. 11, 1–10 (2021).",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.1038/s41598-022-05667-0",
"author": "A Valdés",
"doi-asserted-by": "publisher",
"first-page": "1650",
"journal-title": "Sci. Rep.",
"key": "40999_CR19",
"unstructured": "Valdés, A. et al. Metabolomics study of COVID-19 patients in four different clinical stages. Sci. Rep. 12, 1650 (2022).",
"volume": "12",
"year": "2022"
},
{
"DOI": "10.1038/s41598-021-00755-z",
"author": "M Dei Cas",
"doi-asserted-by": "publisher",
"first-page": "21633",
"journal-title": "Sci. Rep.",
"key": "40999_CR20",
"unstructured": "Dei Cas, M. et al. Link between serum lipid signature and prognostic factors in COVID-19 patients. Sci. Rep. 11, 21633 (2021).",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.3389/fgene.2021.721556",
"author": "MR Hasan",
"doi-asserted-by": "publisher",
"journal-title": "Front. Genet.",
"key": "40999_CR21",
"unstructured": "Hasan, M. R., Suleiman, M. & Pérez-López, A. Metabolomics in the diagnosis and prognosis of COVID-19. Front. Genet. 12, 721556 (2021).",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1038/s41579-022-00846-2",
"author": "HE Davis",
"doi-asserted-by": "publisher",
"first-page": "133",
"journal-title": "Nat. Rev. Microbiol.",
"key": "40999_CR22",
"unstructured": "Davis, H. E., McCorkell, L., Vogel, J. M. & Topol, E. J. Long COVID: Major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 21, 133–146 (2023).",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.3389/fmed.2020.555301",
"author": "RB Aguilar",
"doi-asserted-by": "publisher",
"journal-title": "Front. Med.",
"key": "40999_CR23",
"unstructured": "Aguilar, R. B. et al. Current understanding of COVID-19 clinical course and investigational treatments. Front. Med. 7, 555301 (2020).",
"volume": "7",
"year": "2020"
},
{
"DOI": "10.1021/acs.jproteome.1c00786",
"author": "OE Albóniga",
"doi-asserted-by": "publisher",
"first-page": "623",
"journal-title": "J. Proteome Res.",
"key": "40999_CR24",
"unstructured": "Albóniga, O. E. et al. Metabolic snapshot of plasma samples reveals new pathways implicated in SARS-CoV-2 pathogenesis. J. Proteome Res. 21, 623–634 (2022).",
"volume": "21",
"year": "2022"
},
{
"author": "V Fanelli",
"first-page": "326",
"journal-title": "J. Thorac. Dis.",
"key": "40999_CR25",
"unstructured": "Fanelli, V. et al. Acute respiratory distress syndrome: New definition, current and future therapeutic options. J. Thorac. Dis. 5, 326–334 (2013).",
"volume": "5",
"year": "2013"
},
{
"key": "40999_CR26",
"unstructured": "CDC, Centers for, disease control and prevention, & CDC. Interim Operational Considerations for Public Health Management of Healthcare Workers Exposed to or with Suspected or Confirmed COVID-19: Non-U.S. Healthcare Settings. https://www.cdc.gov/coronavirus/2019-ncov/hcp/non-us-settings/public-health-management-hcwexposed."
},
{
"key": "40999_CR27",
"unstructured": "Lejardi, D. & Technologies, A. Improving Coverage of the Plasma Lipidome Using Iterative MS/MS Data Acquisition Combined with Lipid Annotator Software and 6546 LC/Q-TOF. (2020)."
},
{
"DOI": "10.1039/C5AN01638J",
"author": "J Kuligowski",
"doi-asserted-by": "publisher",
"first-page": "7810",
"journal-title": "Analyst",
"key": "40999_CR28",
"unstructured": "Kuligowski, J., Sánchez-Illana, Á., Sanjuán-Herráez, D., Vento, M. & Quintás, G. Intra-batch effect correction in liquid chromatography-mass spectrometry using quality control samples and support vector regression (QC-SVRC). Analyst 140, 7810–7817 (2015).",
"volume": "140",
"year": "2015"
},
{
"DOI": "10.1021/acs.jproteome.8b00720",
"author": "A Gil-de-la-Fuente",
"doi-asserted-by": "publisher",
"first-page": "797",
"journal-title": "J. Proteome Res.",
"key": "40999_CR29",
"unstructured": "Gil-de-la-Fuente, A. et al. CEU mass mediator 3.0: A metabolite annotation tool. J. Proteome Res. 18, 797–802 (2019).",
"volume": "18",
"year": "2019"
},
{
"DOI": "10.3390/metabo10030101",
"author": "JP Koelmel",
"doi-asserted-by": "publisher",
"first-page": "101",
"journal-title": "Metabolites",
"key": "40999_CR30",
"unstructured": "Koelmel, J. P. et al. Lipid annotator: Towards accurate annotation in non-targeted liquid chromatography high-resolution tandem mass spectrometry (LC-HRMS/MS) lipidomics using a rapid and user-friendly software. Metabolites 10, 101 (2020).",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1038/s41587-020-0531-2",
"author": "H Tsugawa",
"doi-asserted-by": "publisher",
"first-page": "1159",
"journal-title": "Nat. Biotechnol.",
"key": "40999_CR31",
"unstructured": "Tsugawa, H. et al. A lipidome atlas in MS-DIAL 4. Nat. Biotechnol. 38, 1159–1163 (2020).",
"volume": "38",
"year": "2020"
},
{
"DOI": "10.3390/metabo11080488",
"author": "N Köhler",
"doi-asserted-by": "publisher",
"first-page": "488",
"journal-title": "Metabolites",
"key": "40999_CR32",
"unstructured": "Köhler, N., Rose, T. D., Falk, L. & Pauling, J. K. Investigating global lipidome alterations with the lipid network explorer. Metabolites 11, 488 (2021).",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.1016/j.xcrm.2021.100369",
"author": "M Sindelar",
"doi-asserted-by": "publisher",
"journal-title": "Cell Rep. Med.",
"key": "40999_CR33",
"unstructured": "Sindelar, M. et al. Longitudinal metabolomics of human plasma reveals prognostic markers of COVID-19 disease severity. Cell Rep. Med. 2, 100369 (2021).",
"volume": "2",
"year": "2021"
},
{
"DOI": "10.3389/fmolb.2021.737039",
"author": "F Schmelter",
"doi-asserted-by": "publisher",
"journal-title": "Front. Mol. Biosci.",
"key": "40999_CR34",
"unstructured": "Schmelter, F. et al. Metabolic and lipidomic markers differentiate COVID-19 from non-hospitalized and other intensive care patients. Front. Mol. Biosci. 8, 737039 (2021).",
"volume": "8",
"year": "2021"
},
{
"DOI": "10.3390/biom12101488",
"author": "N Žarković",
"doi-asserted-by": "publisher",
"first-page": "1488",
"journal-title": "Biomolecules",
"key": "40999_CR35",
"unstructured": "Žarković, N. et al. Lipidomics revealed plasma phospholipid profile differences between deceased and recovered COVID-19 patients. Biomolecules 12, 1488 (2022).",
"volume": "12",
"year": "2022"
},
{
"DOI": "10.1002/nbm.4637",
"author": "M Bizkarguenaga",
"doi-asserted-by": "publisher",
"first-page": "4637",
"journal-title": "NMR Biomed.",
"key": "40999_CR36",
"unstructured": "Bizkarguenaga, M. et al. Uneven metabolic and lipidomic profiles in recovered COVID-19 patients as investigated by plasma NMR metabolomics. NMR Biomed. 35, 4637 (2022).",
"volume": "35",
"year": "2022"
},
{
"DOI": "10.1007/978-3-030-21735-8_15",
"author": "S Albeituni",
"doi-asserted-by": "publisher",
"first-page": "169",
"key": "40999_CR37",
"unstructured": "Albeituni, S. & Stiban, J. Roles of ceramides and other sphingolipids in immune cell function and inflammation. In The Role of Bioactive Lipids in Cancer, Inflammation and Related Diseases Vol. 1161 (eds Honn, K. V. & Zeldin, D. C.) 169–191 (Springer, 2019).",
"volume-title": "The Role of Bioactive Lipids in Cancer, Inflammation and Related Diseases",
"year": "2019"
},
{
"DOI": "10.1016/j.febslet.2005.05.067",
"author": "A Gómez-Muñoz",
"doi-asserted-by": "publisher",
"first-page": "3744",
"journal-title": "FEBS Lett.",
"key": "40999_CR38",
"unstructured": "Gómez-Muñoz, A. et al. Ceramide-1-phosphate promotes cell survival through activation of the phosphatidylinositol 3-kinase/protein kinase B pathway. FEBS Lett. 579, 3744–3750 (2005).",
"volume": "579",
"year": "2005"
},
{
"DOI": "10.1016/j.bbamem.2015.10.009",
"author": "TY Hou",
"doi-asserted-by": "publisher",
"first-page": "85",
"journal-title": "Biochim. Biophys. Acta BBA Biomembr.",
"key": "40999_CR39",
"unstructured": "Hou, T. Y. et al. n-3 polyunsaturated fatty acids suppress CD4+ T cell proliferation by altering phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2] organization. Biochim. Biophys. Acta BBA Biomembr. 1858, 85–96 (2016).",
"volume": "1858",
"year": "2016"
},
{
"DOI": "10.1038/s41598-021-00286-7",
"author": "MM Khodadoust",
"doi-asserted-by": "publisher",
"first-page": "20866",
"journal-title": "Sci. Rep.",
"key": "40999_CR40",
"unstructured": "Khodadoust, M. M. Inferring a causal relationship between ceramide levels and COVID-19 respiratory distress. Sci. Rep. 11, 20866 (2021).",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.1016/j.mayocp.2021.07.001",
"author": "MP Alexander",
"doi-asserted-by": "publisher",
"first-page": "2561",
"journal-title": "Mayo Clin. Proc.",
"key": "40999_CR41",
"unstructured": "Alexander, M. P. et al. Acute kidney injury in severe COVID-19 has similarities to sepsis-associated kidney injury. Mayo Clin. Proc. 96, 2561–2575 (2021).",
"volume": "96",
"year": "2021"
},
{
"DOI": "10.1038/s41380-021-01309-5",
"author": "J Kornhuber",
"doi-asserted-by": "publisher",
"first-page": "307",
"journal-title": "Mol. Psychiatry",
"key": "40999_CR42",
"unstructured": "Kornhuber, J., Hoertel, N. & Gulbins, E. The acid sphingomyelinase/ceramide system in COVID-19. Mol. Psychiatry 27, 307–314 (2022).",
"volume": "27",
"year": "2022"
},
{
"DOI": "10.1172/jci.insight.156104",
"author": "I Petrache",
"doi-asserted-by": "publisher",
"journal-title": "JCI Insight",
"key": "40999_CR43",
"unstructured": "Petrache, I. et al. Marked elevations in lung and plasma ceramide in COVID-19 linked to microvascular injury. JCI Insight 8, e156104 (2023).",
"volume": "8",
"year": "2023"
},
{
"DOI": "10.3389/fcell.2021.689854",
"author": "K Törnquist",
"doi-asserted-by": "publisher",
"journal-title": "Front. Cell Dev. Biol.",
"key": "40999_CR44",
"unstructured": "Törnquist, K., Asghar, M. Y., Srinivasan, V., Korhonen, L. & Lindholm, D. Sphingolipids as modulators of SARS-CoV-2 infection. Front. Cell Dev. Biol. 9, 689854 (2021).",
"volume": "9",
"year": "2021"
},
{
"DOI": "10.1016/j.cmet.2020.06.016",
"author": "J-W Song",
"doi-asserted-by": "publisher",
"first-page": "188",
"journal-title": "Cell Metab.",
"key": "40999_CR45",
"unstructured": "Song, J.-W. et al. Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell Metab. 32, 188-202.e5 (2020).",
"volume": "32",
"year": "2020"
},
{
"DOI": "10.1016/S2589-7500(22)00112-1",
"author": "SK Byeon",
"doi-asserted-by": "publisher",
"first-page": "e632",
"journal-title": "Lancet Digit. Health",
"key": "40999_CR46",
"unstructured": "Byeon, S. K. et al. Development of a multiomics model for identification of predictive biomarkers for COVID-19 severity: A retrospective cohort study. Lancet Digit. Health 4, e632–e645 (2022).",
"volume": "4",
"year": "2022"
},
{
"DOI": "10.1002/ctm2.1069",
"author": "M Kurano",
"doi-asserted-by": "publisher",
"first-page": "1069",
"journal-title": "Clin. Transl. Med.",
"key": "40999_CR47",
"unstructured": "Kurano, M. et al. Dynamic modulations of sphingolipids and glycerophospholipids in COVID-19. Clin. Transl. Med. 12, 1069 (2022).",
"volume": "12",
"year": "2022"
},
{
"DOI": "10.1136/gutjnl-2020-323826",
"author": "Z Ren",
"doi-asserted-by": "publisher",
"first-page": "1253",
"journal-title": "Gut",
"key": "40999_CR48",
"unstructured": "Ren, Z. et al. Alterations in the human oral and gut microbiomes and lipidomics in COVID-19. Gut 70, 1253–1265 (2021).",
"volume": "70",
"year": "2021"
},
{
"DOI": "10.1038/s41598-021-85788-0",
"author": "JC Páez-Franco",
"doi-asserted-by": "publisher",
"first-page": "6350",
"journal-title": "Sci. Rep.",
"key": "40999_CR49",
"unstructured": "Páez-Franco, J. C. et al. Metabolomics analysis reveals a modified amino acid metabolism that correlates with altered oxygen homeostasis in COVID-19 patients. Sci. Rep. 11, 6350 (2021).",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.2337/dc15-2752",
"author": "J Cobb",
"doi-asserted-by": "publisher",
"first-page": "988",
"journal-title": "Diabetes Care",
"key": "40999_CR50",
"unstructured": "Cobb, J. et al. α-Hydroxybutyric acid is a selective metabolite biomarker of impaired glucose tolerance. Diabetes Care 39, 988–995 (2016).",
"volume": "39",
"year": "2016"
},
{
"DOI": "10.1016/j.plipres.2021.101092",
"author": "I Casari",
"doi-asserted-by": "publisher",
"journal-title": "Prog. Lipid Res.",
"key": "40999_CR51",
"unstructured": "Casari, I., Manfredi, M., Metharom, P. & Falasca, M. Dissecting lipid metabolism alterations in SARS-CoV-2. Prog. Lipid Res. 82, 101092 (2021).",
"volume": "82",
"year": "2021"
},
{
"DOI": "10.1016/j.metabol.2021.154739",
"author": "D Shi",
"doi-asserted-by": "publisher",
"journal-title": "Metabolism",
"key": "40999_CR52",
"unstructured": "Shi, D. et al. The serum metabolome of COVID-19 patients is distinctive and predictive. Metabolism 118, 154739 (2021).",
"volume": "118",
"year": "2021"
},
{
"DOI": "10.5501/wjv.v11.i5.237",
"author": "R Kumar",
"doi-asserted-by": "publisher",
"first-page": "237",
"journal-title": "World J. Virol.",
"key": "40999_CR53",
"unstructured": "Kumar, R., Kumar, V., Arya, R., Anand, U. & Priyadarshi, R. N. Association of COVID-19 with hepatic metabolic dysfunction. World J. Virol. 11, 237–251 (2022).",
"volume": "11",
"year": "2022"
},
{
"DOI": "10.3389/fimmu.2021.689966",
"author": "J Torres-Ruiz",
"doi-asserted-by": "publisher",
"journal-title": "Front. Immunol.",
"key": "40999_CR54",
"unstructured": "Torres-Ruiz, J. et al. Redefining COVID-19 severity and prognosis: The role of clinical and immunobiotypes. Front. Immunol. 12, 689966 (2021).",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.3390/ijms21228623",
"author": "E Barberis",
"doi-asserted-by": "publisher",
"first-page": "8623",
"journal-title": "Int. J. Mol. Sci.",
"key": "40999_CR55",
"unstructured": "Barberis, E. et al. Large-scale plasma analysis revealed new mechanisms and molecules associated with the host response to SARS-CoV-2. Int. J. Mol. Sci. 21, 8623 (2020).",
"volume": "21",
"year": "2020"
},
{
"DOI": "10.3390/ijms21228623",
"author": "E Barberis",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Int. J. Mol. Sci.",
"key": "40999_CR56",
"unstructured": "Barberis, E. et al. Large-scale plasma analysis revealed new mechanisms and molecules associated with the host response to sars-cov-2. Int. J. Mol. Sci. 21, 1–25 (2020).",
"volume": "21",
"year": "2020"
},
{
"DOI": "10.3390/ijms221910198",
"author": "E Torretta",
"doi-asserted-by": "publisher",
"first-page": "10198",
"journal-title": "Int. J. Mol. Sci.",
"key": "40999_CR57",
"unstructured": "Torretta, E. et al. Severity of COVID-19 patients predicted by serum sphingolipids signature. Int. J. Mol. Sci. 22, 10198 (2021).",
"volume": "22",
"year": "2021"
},
{
"key": "40999_CR58",
"unstructured": "HMDB. Human Metabolome Database: Showing metabocard for Aminomalonic acid (HMDB0001147). https://hmdb.ca/metabolites/HMDB0001147."
},
{
"DOI": "10.1111/j.1745-4565.1982.tb00429.x",
"author": "JC Hierholzer",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "J. Food Saf.",
"key": "40999_CR59",
"unstructured": "Hierholzer, J. C. & Kabara, J. J. In vitro effects of monolaurin compounds on enveloped RNA and DNA viruses. J. Food Saf. 4, 1–12 (1982).",
"volume": "4",
"year": "1982"
},
{
"DOI": "10.1038/s41586-021-03237-4",
"author": "BA Johnson",
"doi-asserted-by": "publisher",
"first-page": "293",
"journal-title": "Nature",
"key": "40999_CR60",
"unstructured": "Johnson, B. A. et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 591, 293–299 (2021).",
"volume": "591",
"year": "2021"
},
{
"DOI": "10.1128/jvi.00128-22",
"author": "R Essalmani",
"doi-asserted-by": "publisher",
"journal-title": "J. Virol.",
"key": "40999_CR61",
"unstructured": "Essalmani, R. et al. Distinctive roles of furin and TMPRSS2 in SARS-CoV-2 infectivity. J. Virol. 96, e0012822 (2022).",
"volume": "96",
"year": "2022"
},
{
"DOI": "10.1016/j.aca.2021.339405",
"author": "A Gomez-Gomez",
"doi-asserted-by": "publisher",
"journal-title": "Anal. Chim. Acta",
"key": "40999_CR62",
"unstructured": "Gomez-Gomez, A. et al. Untargeted detection of the carbonyl metabolome by chemical derivatization and liquid chromatography-tandem mass spectrometry in precursor ion scan mode: Elucidation of COVID-19 severity biomarkers. Anal. Chim. Acta 1196, 339405 (2022).",
"volume": "1196",
"year": "2022"
},
{
"key": "40999_CR63",
"unstructured": "HMDB. Human Metabolome Database: Showing Metabocard for Threonic acid (HMDB0000943). https://hmdb.ca/metabolites/HMDB0000943."
},
{
"DOI": "10.3390/nu13041172",
"author": "GP Milani",
"doi-asserted-by": "publisher",
"first-page": "1172",
"journal-title": "Nutrients",
"key": "40999_CR64",
"unstructured": "Milani, G. P., Macchi, M. & Guz-Mark, A. Vitamin C in the treatment of COVID-19. Nutrients 13, 1172 (2021).",
"volume": "13",
"year": "2021"
},
{
"DOI": "10.1097/MD.0000000000026427",
"author": "GAS Toscano",
"doi-asserted-by": "publisher",
"first-page": "e26427",
"journal-title": "Medicine",
"key": "40999_CR65",
"unstructured": "Toscano, G. A. S., de Araújo, I. I., de Souza, T. A., Barbosa Mirabal, I. R. & de Vasconcelos Torres, G. Vitamin C and D supplementation and the severity of COVID-19: A protocol for systematic review and meta-analysis. Medicine 100, e26427 (2021).",
"volume": "100",
"year": "2021"
},
{
"author": "E Kočar",
"first-page": "158849",
"journal-title": "Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids",
"key": "40999_CR66",
"unstructured": "Kočar, E., Režen, T. & Rozman, D. Cholesterol, lipoproteins, and COVID-19: Basic concepts and clinical applications. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 1866, 158849 (2021).",
"volume": "1866",
"year": "2021"
},
{
"DOI": "10.1248/bpb.29.1538",
"author": "T Suzuki",
"doi-asserted-by": "publisher",
"first-page": "1538",
"journal-title": "Biol. Pharm. Bull.",
"key": "40999_CR67",
"unstructured": "Suzuki, T. & Suzuki, Y. Virus infection and lipid rafts. Biol. Pharm. Bull. 29, 1538–1541 (2006).",
"volume": "29",
"year": "2006"
}
],
"reference-count": 67,
"references-count": 67,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.nature.com/articles/s41598-023-40999-5"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Multidisciplinary"
],
"subtitle": [],
"title": "Differential abundance of lipids and metabolites related to SARS-CoV-2 infection and susceptibility",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1007/springer_crossmark_policy",
"volume": "13"
}